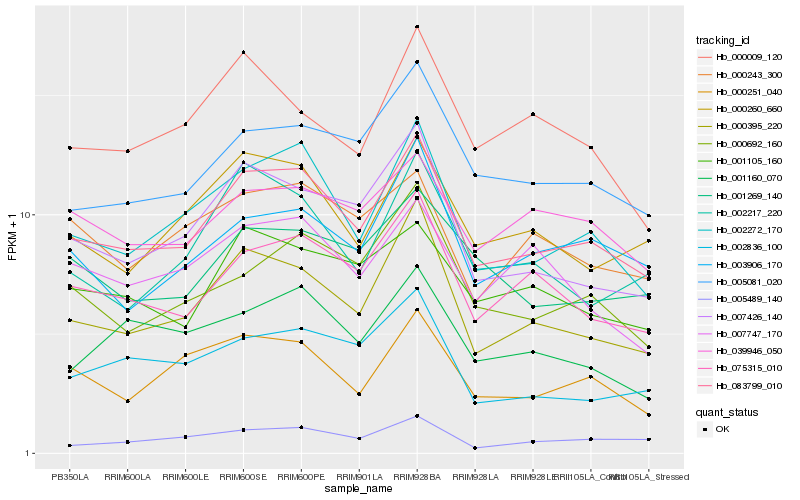
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_083799_010 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_002272_170 |
0.0654948896 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase-like protein 1 [Jatropha curcas] |
| 3 |
Hb_000395_220 |
0.0729443539 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 4 |
Hb_075315_010 |
0.0774919105 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 5 |
Hb_005081_020 |
0.0818550334 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 6 |
Hb_007426_140 |
0.0840751781 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 7 |
Hb_000260_660 |
0.087006569 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001105_160 |
0.0893611934 |
- |
- |
PREDICTED: protein phosphatase 2C 32 [Jatropha curcas] |
| 9 |
Hb_003906_170 |
0.0946481254 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 10 |
Hb_000692_160 |
0.0959233629 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 11 |
Hb_039946_050 |
0.0961108797 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_000251_040 |
0.0973058526 |
- |
- |
PREDICTED: DNA polymerase alpha catalytic subunit isoform X1 [Jatropha curcas] |
| 13 |
Hb_001269_140 |
0.0977456117 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 14 |
Hb_000243_300 |
0.098938127 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_005489_140 |
0.1003532274 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 16 |
Hb_002217_220 |
0.1016792021 |
- |
- |
hypothetical protein POPTR_0010s15930g [Populus trichocarpa] |
| 17 |
Hb_002836_100 |
0.1023966856 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007747_170 |
0.103929134 |
- |
- |
PREDICTED: uncharacterized protein At5g41620 [Jatropha curcas] |
| 19 |
Hb_001160_070 |
0.10400741 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 20 |
Hb_000009_120 |
0.1068500257 |
- |
- |
PREDICTED: probable Xaa-Pro aminopeptidase P isoform X1 [Jatropha curcas] |