| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
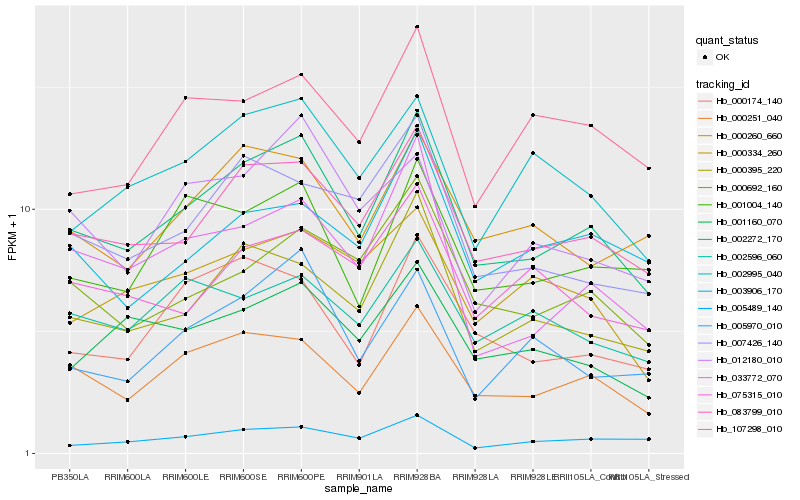
Hb_002272_170 |
0.0 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase-like protein 1 [Jatropha curcas] |
| 2 |
Hb_083799_010 |
0.0654948896 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 3 |
Hb_000251_040 |
0.0758835919 |
- |
- |
PREDICTED: DNA polymerase alpha catalytic subunit isoform X1 [Jatropha curcas] |
| 4 |
Hb_000395_220 |
0.0915479545 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 5 |
Hb_000692_160 |
0.1042176389 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 6 |
Hb_033772_070 |
0.1045055697 |
- |
- |
PREDICTED: uncharacterized protein LOC105633681 [Jatropha curcas] |
| 7 |
Hb_007426_140 |
0.1068812207 |
- |
- |
ornithine aminotransferase, putative [Ricinus communis] |
| 8 |
Hb_001160_070 |
0.1077274471 |
- |
- |
PREDICTED: uncharacterized protein LOC105632163 [Jatropha curcas] |
| 9 |
Hb_000260_660 |
0.1084806852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001004_140 |
0.1092788864 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 11 |
Hb_005489_140 |
0.1112997747 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 12 |
Hb_002995_040 |
0.1124233271 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase dyrk1 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000174_140 |
0.1131366242 |
- |
- |
JHL23J11.8 [Jatropha curcas] |
| 14 |
Hb_000334_260 |
0.1167387905 |
- |
- |
carotenoid cleavage dioxygenase 1 [Manihot esculenta] |
| 15 |
Hb_005970_010 |
0.1181777249 |
- |
- |
PREDICTED: probable isoprenylcysteine alpha-carbonyl methylesterase ICMEL1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_002596_060 |
0.1186761851 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_075315_010 |
0.1196217656 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |
| 18 |
Hb_003906_170 |
0.1203917727 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 19 |
Hb_107298_010 |
0.1207384633 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF2A isoform X2 [Jatropha curcas] |
| 20 |
Hb_012180_010 |
0.1213287807 |
- |
- |
PREDICTED: (S)-ureidoglycine aminohydrolase [Jatropha curcas] |