| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
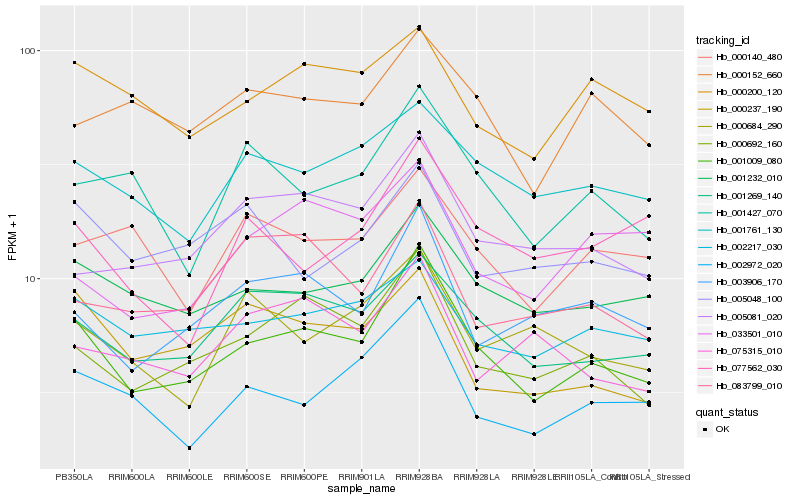
Hb_001009_080 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000692_160 |
0.0963069335 |
- |
- |
Transportin 1 isoform 1 [Theobroma cacao] |
| 3 |
Hb_001269_140 |
0.1055250664 |
- |
- |
PREDICTED: nuclear pore complex protein NUP96 [Jatropha curcas] |
| 4 |
Hb_001232_010 |
0.1069438573 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 5 |
Hb_003906_170 |
0.117307004 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_002972_020 |
0.1199823589 |
- |
- |
PREDICTED: probable ubiquitin-like-specific protease 2A isoform X1 [Jatropha curcas] |
| 7 |
Hb_001761_130 |
0.1252821728 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002217_030 |
0.1268413806 |
- |
- |
PREDICTED: DNA (cytosine-5)-methyltransferase DRM2 isoform X1 [Jatropha curcas] |
| 9 |
Hb_005081_020 |
0.1300777807 |
- |
- |
PREDICTED: ninja-family protein mc410 [Jatropha curcas] |
| 10 |
Hb_001427_070 |
0.1309691369 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 11 |
Hb_000200_120 |
0.1360515192 |
- |
- |
PREDICTED: probable fructose-bisphosphate aldolase 3, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000152_660 |
0.1379108402 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 13 |
Hb_000237_190 |
0.1402063814 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000140_480 |
0.1427104382 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 15 |
Hb_077562_030 |
0.1431912954 |
- |
- |
PREDICTED: cytochrome P450 4X1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_083799_010 |
0.1440462969 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 17 |
Hb_005048_100 |
0.1444417683 |
- |
- |
secretory carrier membrane protein, putative [Ricinus communis] |
| 18 |
Hb_000684_290 |
0.1445967405 |
- |
- |
ubiquitin protein ligase E3a, putative [Ricinus communis] |
| 19 |
Hb_033501_010 |
0.1459705373 |
- |
- |
hypothetical protein JCGZ_21381 [Jatropha curcas] |
| 20 |
Hb_075315_010 |
0.1462857821 |
- |
- |
ornithine aminotransferase [Camellia sinensis] |