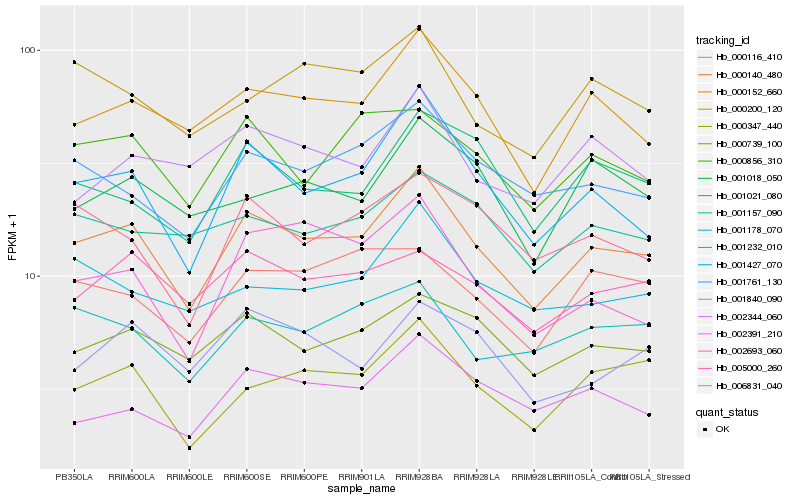
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000140_480 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 2 |
Hb_001427_070 |
0.0711553875 |
- |
- |
PREDICTED: xaa-Pro dipeptidase [Jatropha curcas] |
| 3 |
Hb_000152_660 |
0.0758571249 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 4 |
Hb_000739_100 |
0.089128687 |
- |
- |
ku P80 DNA helicase, putative [Ricinus communis] |
| 5 |
Hb_001761_130 |
0.0893496673 |
- |
- |
PREDICTED: uncharacterized protein LOC105646118 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000347_440 |
0.0917811756 |
transcription factor |
TF Family: GRAS |
PREDICTED: nodulation-signaling pathway 1 protein [Jatropha curcas] |
| 7 |
Hb_000856_310 |
0.0963447928 |
- |
- |
PREDICTED: hypersensitive-induced response protein 1 [Jatropha curcas] |
| 8 |
Hb_006831_040 |
0.0977738417 |
- |
- |
PREDICTED: enhancer of mRNA-decapping protein 4-like isoform X1 [Jatropha curcas] |
| 9 |
Hb_001232_010 |
0.0993529501 |
- |
- |
PREDICTED: uncharacterized protein LOC105634976 [Jatropha curcas] |
| 10 |
Hb_001178_070 |
0.0996487053 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein FBL11 [Jatropha curcas] |
| 11 |
Hb_001021_080 |
0.0998749288 |
- |
- |
PREDICTED: autophagy-related protein 18a [Jatropha curcas] |
| 12 |
Hb_002391_210 |
0.1000871929 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 13 |
Hb_005000_260 |
0.1001882341 |
- |
- |
recA family protein [Populus trichocarpa] |
| 14 |
Hb_000200_120 |
0.1025611958 |
- |
- |
PREDICTED: probable fructose-bisphosphate aldolase 3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_001157_090 |
0.103322502 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase SINAT2-like [Jatropha curcas] |
| 16 |
Hb_000116_410 |
0.1042371454 |
- |
- |
hypothetical protein JCGZ_22416 [Jatropha curcas] |
| 17 |
Hb_001018_050 |
0.1048846556 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 18 |
Hb_002344_060 |
0.1053107386 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 19 |
Hb_001840_090 |
0.106250229 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 20 |
Hb_002693_060 |
0.1063189221 |
- |
- |
Protein kinase APK1B, chloroplast precursor, putative [Ricinus communis] |