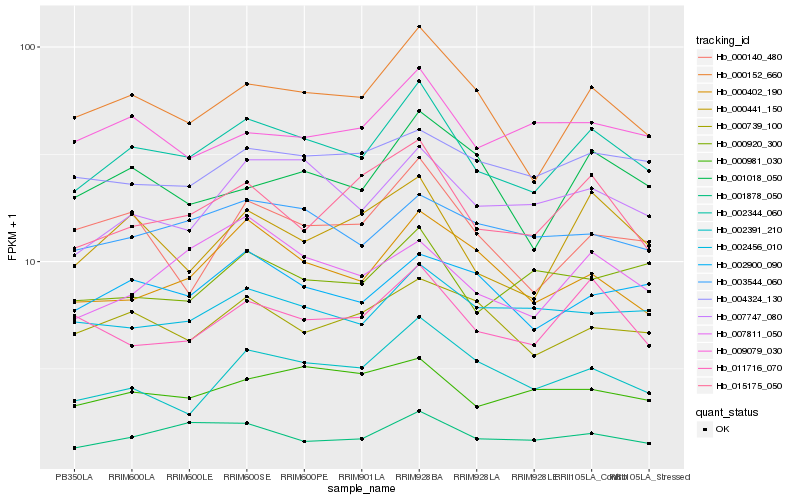
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002344_060 |
0.0 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 2 |
Hb_000152_660 |
0.0752610962 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 3 |
Hb_001878_050 |
0.086836177 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 4 |
Hb_007747_080 |
0.0873098974 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000441_150 |
0.0893417253 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_015175_050 |
0.0905895464 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10 isoform X3 [Jatropha curcas] |
| 7 |
Hb_011716_070 |
0.0949699453 |
transcription factor |
TF Family: HRT |
PREDICTED: uncharacterized protein LOC105637858 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002456_010 |
0.0978727438 |
- |
- |
PREDICTED: protein kinase and PP2C-like domain-containing protein isoform X2 [Jatropha curcas] |
| 9 |
Hb_003544_060 |
0.0982242949 |
- |
- |
hypothetical protein JCGZ_22100 [Jatropha curcas] |
| 10 |
Hb_000402_190 |
0.0988564843 |
- |
- |
PREDICTED: F-box protein At1g70590 [Jatropha curcas] |
| 11 |
Hb_002900_090 |
0.0989449321 |
transcription factor |
TF Family: MYB |
PREDICTED: uncharacterized protein LOC105646817 [Jatropha curcas] |
| 12 |
Hb_000981_030 |
0.0999036963 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 13 |
Hb_002391_210 |
0.1009042154 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 14 |
Hb_001018_050 |
0.1012853539 |
- |
- |
PREDICTED: aspartate aminotransferase, cytoplasmic [Jatropha curcas] |
| 15 |
Hb_007811_050 |
0.1016604248 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase NAK [Jatropha curcas] |
| 16 |
Hb_000739_100 |
0.1016887886 |
- |
- |
ku P80 DNA helicase, putative [Ricinus communis] |
| 17 |
Hb_009079_030 |
0.1025407239 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 18 |
Hb_004324_130 |
0.1049095189 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 19 |
Hb_000140_480 |
0.1053107386 |
- |
- |
PREDICTED: uncharacterized protein LOC105637350 [Jatropha curcas] |
| 20 |
Hb_000920_300 |
0.1060746074 |
- |
- |
PREDICTED: flowering time control protein FY isoform X2 [Jatropha curcas] |