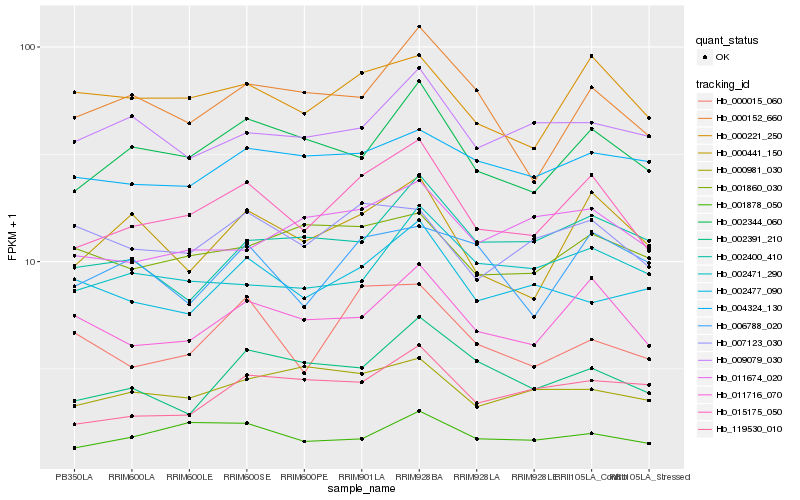
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_015175_050 |
0.0 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 10 isoform X3 [Jatropha curcas] |
| 2 |
Hb_002344_060 |
0.0905895464 |
- |
- |
PREDICTED: methylthioribose-1-phosphate isomerase [Jatropha curcas] |
| 3 |
Hb_011716_070 |
0.0910852307 |
transcription factor |
TF Family: HRT |
PREDICTED: uncharacterized protein LOC105637858 isoform X1 [Jatropha curcas] |
| 4 |
Hb_000221_250 |
0.0952807636 |
- |
- |
hypothetical protein JCGZ_15814 [Jatropha curcas] |
| 5 |
Hb_000441_150 |
0.1005763695 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_001878_050 |
0.1072371248 |
- |
- |
PREDICTED: uncharacterized protein LOC105646110 isoform X3 [Jatropha curcas] |
| 7 |
Hb_000152_660 |
0.1108589073 |
- |
- |
PREDICTED: BAG family molecular chaperone regulator 7 [Jatropha curcas] |
| 8 |
Hb_000015_060 |
0.11177142 |
- |
- |
PREDICTED: uncharacterized protein LOC104239844 isoform X2 [Nicotiana sylvestris] |
| 9 |
Hb_007123_030 |
0.1158421992 |
- |
- |
PREDICTED: uncharacterized protein LOC105634056 isoform X3 [Jatropha curcas] |
| 10 |
Hb_011674_020 |
0.1174252459 |
- |
- |
PREDICTED: NEDD8-activating enzyme E1 regulatory subunit-like [Jatropha curcas] |
| 11 |
Hb_002391_210 |
0.1182250531 |
- |
- |
PREDICTED: putative F-box protein At1g65770 [Jatropha curcas] |
| 12 |
Hb_002471_290 |
0.1205340778 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_119530_010 |
0.120603212 |
- |
- |
- |
| 14 |
Hb_009079_030 |
0.1209522716 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 15 |
Hb_002477_090 |
0.1216826971 |
- |
- |
PREDICTED: uncharacterized protein LOC105631409 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000981_030 |
0.1218338233 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105640649 [Jatropha curcas] |
| 17 |
Hb_004324_130 |
0.1222021875 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 18 |
Hb_001860_030 |
0.1224347592 |
- |
- |
PREDICTED: cleavage stimulation factor subunit 77 isoform X5 [Jatropha curcas] |
| 19 |
Hb_006788_020 |
0.1229947748 |
- |
- |
hypothetical protein JCGZ_21864 [Jatropha curcas] |
| 20 |
Hb_002400_410 |
0.1232051418 |
- |
- |
PREDICTED: topless-related protein 1-like [Jatropha curcas] |