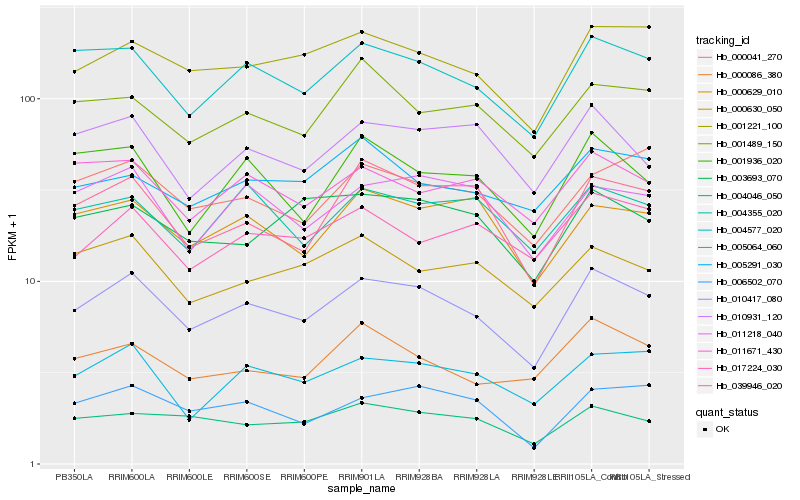
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010417_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645742 isoform X1 [Jatropha curcas] |
| 2 |
Hb_004577_020 |
0.0613199632 |
- |
- |
PREDICTED: MOB kinase activator-like 1 [Jatropha curcas] |
| 3 |
Hb_001221_100 |
0.0753404479 |
- |
- |
PREDICTED: elongation factor 1-delta 1-like [Citrus sinensis] |
| 4 |
Hb_005064_060 |
0.0858319851 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_004046_050 |
0.0866065224 |
- |
- |
PREDICTED: origin of replication complex subunit 3 [Jatropha curcas] |
| 6 |
Hb_011218_040 |
0.0866936073 |
- |
- |
methylenetetrahydrofolate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_039946_020 |
0.0873926099 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006502_070 |
0.0898790228 |
- |
- |
separase, putative [Ricinus communis] |
| 9 |
Hb_000629_010 |
0.0898897928 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 32 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001936_020 |
0.09241358 |
- |
- |
PREDICTED: phosphatidylinositol 3-kinase, root isoform isoform X1 [Jatropha curcas] |
| 11 |
Hb_000086_380 |
0.0925460519 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 12 |
Hb_011671_430 |
0.0928924641 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |
| 13 |
Hb_000041_270 |
0.0930512643 |
- |
- |
PREDICTED: mitochondrial import receptor subunit TOM40-1 [Jatropha curcas] |
| 14 |
Hb_010931_120 |
0.0931736351 |
- |
- |
myosin heavy chain-related family protein [Populus trichocarpa] |
| 15 |
Hb_000630_050 |
0.0937171949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001489_150 |
0.094535116 |
- |
- |
PREDICTED: uncharacterized protein LOC105123623 isoform X2 [Populus euphratica] |
| 17 |
Hb_004355_020 |
0.0952561071 |
- |
- |
PREDICTED: lipoamide acyltransferase component of branched-chain alpha-keto acid dehydrogenase complex, mitochondrial [Jatropha curcas] |
| 18 |
Hb_017224_030 |
0.0956123101 |
- |
- |
PREDICTED: A-agglutinin anchorage subunit [Jatropha curcas] |
| 19 |
Hb_003693_070 |
0.0958915692 |
- |
- |
PREDICTED: S-(hydroxymethyl)glutathione dehydrogenase [Jatropha curcas] |
| 20 |
Hb_005291_030 |
0.0966493694 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |