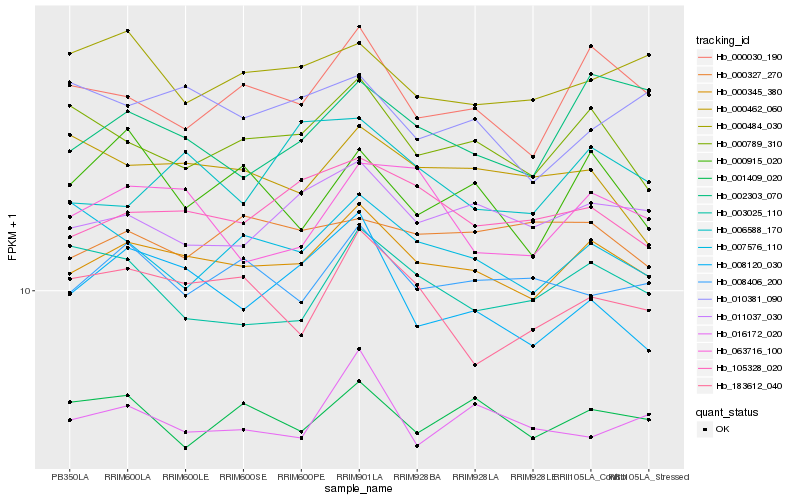
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000345_380 |
0.0 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 7, peroxisomal [Jatropha curcas] |
| 2 |
Hb_105328_020 |
0.0522727664 |
- |
- |
PREDICTED: phosphatidylinositol 3,4,5-trisphosphate 3-phosphatase and dual-specificity protein phosphatase PTEN [Jatropha curcas] |
| 3 |
Hb_011037_030 |
0.0538246737 |
- |
- |
PREDICTED: uncharacterized protein LOC105641407 [Jatropha curcas] |
| 4 |
Hb_000030_190 |
0.0540564922 |
- |
- |
PREDICTED: TATA-box-binding protein-like isoform X1 [Citrus sinensis] |
| 5 |
Hb_000789_310 |
0.0581543076 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PRT1 [Jatropha curcas] |
| 6 |
Hb_008406_200 |
0.0586098561 |
transcription factor |
TF Family: SNF2 |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_001409_020 |
0.0598832469 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105638648 [Jatropha curcas] |
| 8 |
Hb_016172_020 |
0.0645430456 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 9 |
Hb_063716_100 |
0.0647887695 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 10 |
Hb_002303_070 |
0.0658954498 |
- |
- |
PREDICTED: uncharacterized RNA-binding protein C1827.05c [Jatropha curcas] |
| 11 |
Hb_000484_030 |
0.0664211335 |
- |
- |
PREDICTED: probable 26S proteasome non-ATPase regulatory subunit 3 [Jatropha curcas] |
| 12 |
Hb_000327_270 |
0.0674383914 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_007576_110 |
0.0675494147 |
- |
- |
Endoplasmic reticulum vesicle transporter protein [Theobroma cacao] |
| 14 |
Hb_183612_040 |
0.0682698612 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 15 |
Hb_010381_090 |
0.069343279 |
- |
- |
PREDICTED: protein FLX-like 3 [Jatropha curcas] |
| 16 |
Hb_008120_030 |
0.0695367741 |
- |
- |
PREDICTED: uncharacterized protein LOC101216675 [Cucumis sativus] |
| 17 |
Hb_000462_060 |
0.0697614187 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000915_020 |
0.0705617558 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_006588_170 |
0.0707260559 |
- |
- |
PREDICTED: OTU domain-containing protein At3g57810 [Jatropha curcas] |
| 20 |
Hb_003025_110 |
0.0709653499 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |