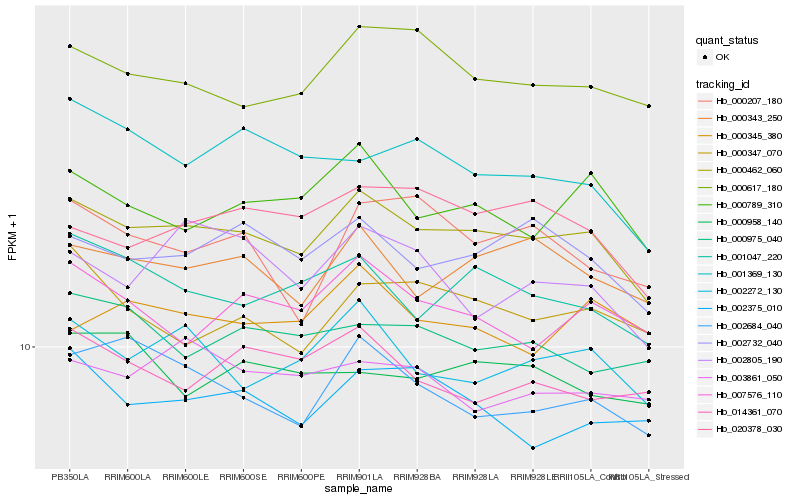
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000462_060 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002732_040 |
0.0533741153 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000343_250 |
0.0537292213 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 4 |
Hb_002272_130 |
0.0546644661 |
- |
- |
PREDICTED: DNA excision repair protein ERCC-1 [Jatropha curcas] |
| 5 |
Hb_000789_310 |
0.0565130537 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase PRT1 [Jatropha curcas] |
| 6 |
Hb_000617_180 |
0.0565573579 |
- |
- |
hypothetical protein B456_013G125900 [Gossypium raimondii] |
| 7 |
Hb_001047_220 |
0.0575834105 |
- |
- |
PREDICTED: F-box protein SKIP17-like [Jatropha curcas] |
| 8 |
Hb_020378_030 |
0.0624218855 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 9 |
Hb_007576_110 |
0.0626727561 |
- |
- |
Endoplasmic reticulum vesicle transporter protein [Theobroma cacao] |
| 10 |
Hb_002684_040 |
0.0633194412 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000347_070 |
0.066772691 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002375_010 |
0.067880111 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000975_040 |
0.0679665944 |
- |
- |
hypothetical protein CICLE_v10028249mg [Citrus clementina] |
| 14 |
Hb_003861_050 |
0.0681992285 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 15 |
Hb_014361_070 |
0.0696726442 |
- |
- |
PREDICTED: molybdate-anion transporter [Jatropha curcas] |
| 16 |
Hb_000345_380 |
0.0697614187 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 7, peroxisomal [Jatropha curcas] |
| 17 |
Hb_001369_130 |
0.0698572888 |
- |
- |
PREDICTED: autophagy-related protein 3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000207_180 |
0.0701970844 |
- |
- |
PREDICTED: calponin homology domain-containing protein DDB_G0272472 [Jatropha curcas] |
| 19 |
Hb_002805_190 |
0.0708647843 |
- |
- |
spliceosome associated protein, putative [Ricinus communis] |
| 20 |
Hb_000958_140 |
0.0709245921 |
- |
- |
PREDICTED: uncharacterized protein LOC105628714 [Jatropha curcas] |