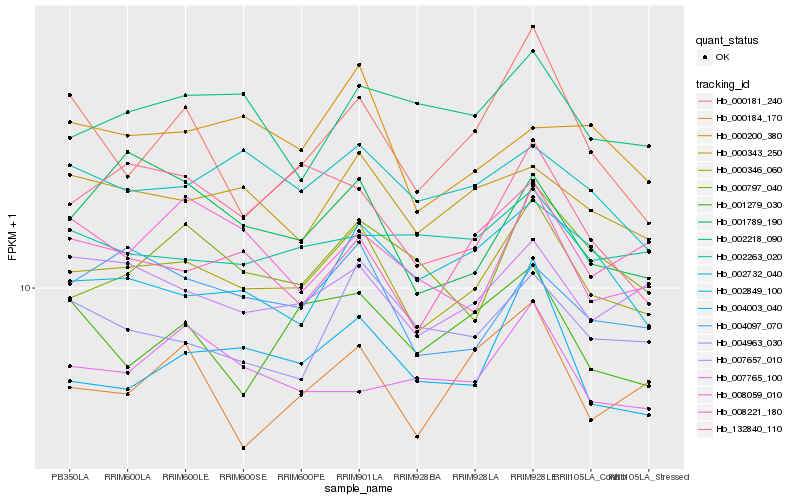
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000346_060 |
0.0 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 2 |
Hb_007657_010 |
0.0600691854 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 3 |
Hb_000343_250 |
0.061447596 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 4 |
Hb_004003_040 |
0.0629592957 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 5 |
Hb_004097_070 |
0.0638410221 |
- |
- |
PREDICTED: F-box protein At2g26160-like [Jatropha curcas] |
| 6 |
Hb_002732_040 |
0.0687451314 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004963_030 |
0.0709245025 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Jatropha curcas] |
| 8 |
Hb_000797_040 |
0.0722402366 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 9 |
Hb_001789_190 |
0.0732509001 |
- |
- |
PREDICTED: uncharacterized protein LOC105648441 [Jatropha curcas] |
| 10 |
Hb_002849_100 |
0.0735180055 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 1, mitochondrial [Jatropha curcas] |
| 11 |
Hb_008059_010 |
0.074158624 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000184_170 |
0.0743157381 |
- |
- |
hypothetical protein RCOM_1213430 [Ricinus communis] |
| 13 |
Hb_008221_180 |
0.075135381 |
- |
- |
PREDICTED: uncharacterized protein LOC105644223 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002218_090 |
0.0754850006 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 15 |
Hb_000200_380 |
0.0756260066 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X2 [Jatropha curcas] |
| 16 |
Hb_132840_110 |
0.0760839106 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 17 |
Hb_002263_020 |
0.0767949572 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 18 |
Hb_001279_030 |
0.0784642146 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A 57 kDa regulatory subunit B' theta isoform-like [Jatropha curcas] |
| 19 |
Hb_000181_240 |
0.0789389039 |
- |
- |
PREDICTED: uncharacterized protein LOC105637672 [Jatropha curcas] |
| 20 |
Hb_007765_100 |
0.079495794 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |