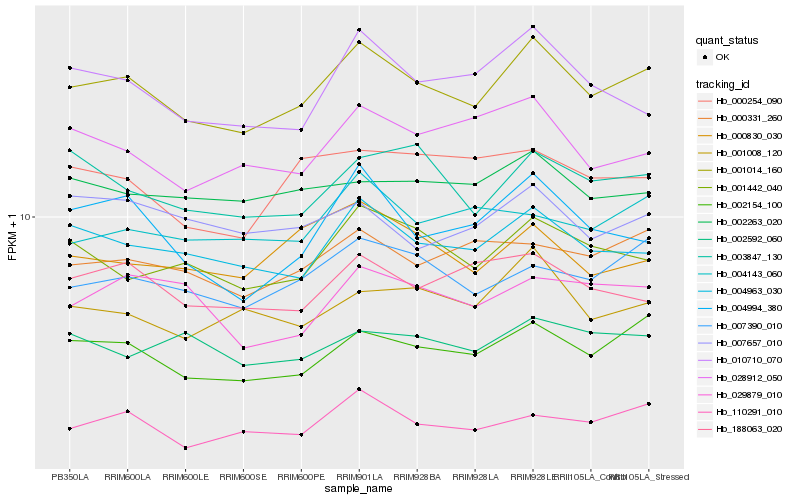
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001014_160 |
0.0 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 2 |
Hb_002154_100 |
0.053007337 |
- |
- |
PREDICTED: putative ATP-dependent RNA helicase DHX33 [Jatropha curcas] |
| 3 |
Hb_004963_030 |
0.0591776005 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Jatropha curcas] |
| 4 |
Hb_007390_010 |
0.0675066022 |
- |
- |
catalytic, putative [Ricinus communis] |
| 5 |
Hb_001442_040 |
0.0706521728 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000331_260 |
0.0717987299 |
- |
- |
PREDICTED: WD repeat-containing protein 74 isoform X2 [Jatropha curcas] |
| 7 |
Hb_110291_010 |
0.0726768029 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_010710_070 |
0.0729654879 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 9 |
Hb_001008_120 |
0.0738883072 |
desease resistance |
Gene Name: PEX-1N |
peroxisome biogenesis factor, putative [Ricinus communis] |
| 10 |
Hb_003847_130 |
0.0752935704 |
- |
- |
cytochrome C oxidase assembly protein cox11, putative [Ricinus communis] |
| 11 |
Hb_028912_050 |
0.0756570565 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 12 |
Hb_004994_380 |
0.0760779794 |
- |
- |
hypothetical protein JCGZ_14410 [Jatropha curcas] |
| 13 |
Hb_188063_020 |
0.0768135278 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000830_030 |
0.0780309159 |
- |
- |
PREDICTED: uncharacterized protein LOC105644214 [Jatropha curcas] |
| 15 |
Hb_002263_020 |
0.0780430961 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 16 |
Hb_004143_060 |
0.0802303166 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_029879_010 |
0.08053989 |
- |
- |
ribosomal pseudouridine synthase, putative [Ricinus communis] |
| 18 |
Hb_007657_010 |
0.0811107447 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 19 |
Hb_002592_060 |
0.0811962367 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000254_090 |
0.0822795918 |
- |
- |
PREDICTED: la protein 2 [Jatropha curcas] |