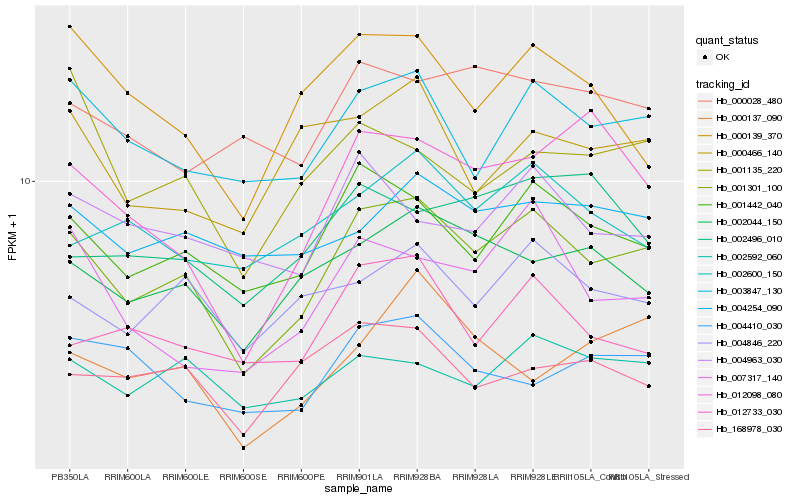
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001301_100 |
0.0 |
- |
- |
transporter, putative [Ricinus communis] |
| 2 |
Hb_001442_040 |
0.0640872978 |
- |
- |
PREDICTED: peroxisome biogenesis factor 10 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003847_130 |
0.0739706194 |
- |
- |
cytochrome C oxidase assembly protein cox11, putative [Ricinus communis] |
| 4 |
Hb_002044_150 |
0.078123377 |
- |
- |
PREDICTED: A/G-specific adenine DNA glycosylase isoform X1 [Jatropha curcas] |
| 5 |
Hb_004846_220 |
0.0803010242 |
- |
- |
PREDICTED: probable protein phosphatase 2C 11 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007317_140 |
0.0823699373 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 7 |
Hb_002592_060 |
0.0861277791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_012733_030 |
0.0878369772 |
- |
- |
hypothetical protein OsI_15243 [Oryza sativa Indica Group] |
| 9 |
Hb_004254_090 |
0.0883562656 |
- |
- |
PREDICTED: cleavage stimulating factor 64 [Jatropha curcas] |
| 10 |
Hb_012098_080 |
0.0897197428 |
- |
- |
KOM, putative [Ricinus communis] |
| 11 |
Hb_004963_030 |
0.0905839433 |
- |
- |
PREDICTED: twinkle homolog protein, chloroplastic/mitochondrial [Jatropha curcas] |
| 12 |
Hb_000466_140 |
0.0909444496 |
- |
- |
hypothetical protein JCGZ_02122 [Jatropha curcas] |
| 13 |
Hb_002600_150 |
0.0911426233 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |
| 14 |
Hb_168978_030 |
0.0912015354 |
- |
- |
PREDICTED: CBL-interacting serine/threonine-protein kinase 8 [Jatropha curcas] |
| 15 |
Hb_000137_090 |
0.0921346582 |
- |
- |
PREDICTED: HAUS augmin-like complex subunit 3 [Vitis vinifera] |
| 16 |
Hb_000028_480 |
0.0921410699 |
- |
- |
PREDICTED: putative 3,4-dihydroxy-2-butanone kinase [Jatropha curcas] |
| 17 |
Hb_000139_370 |
0.0923003452 |
- |
- |
ubiquitin-activating enzyme E1c, putative [Ricinus communis] |
| 18 |
Hb_001135_220 |
0.0932741945 |
- |
- |
FGFR1 oncogene partner, putative [Ricinus communis] |
| 19 |
Hb_002496_010 |
0.0934381781 |
- |
- |
PREDICTED: protein GDAP2 homolog isoform X1 [Jatropha curcas] |
| 20 |
Hb_004410_030 |
0.0943914857 |
- |
- |
PREDICTED: protein ACCUMULATION AND REPLICATION OF CHLOROPLASTS 6, chloroplastic isoform X1 [Jatropha curcas] |