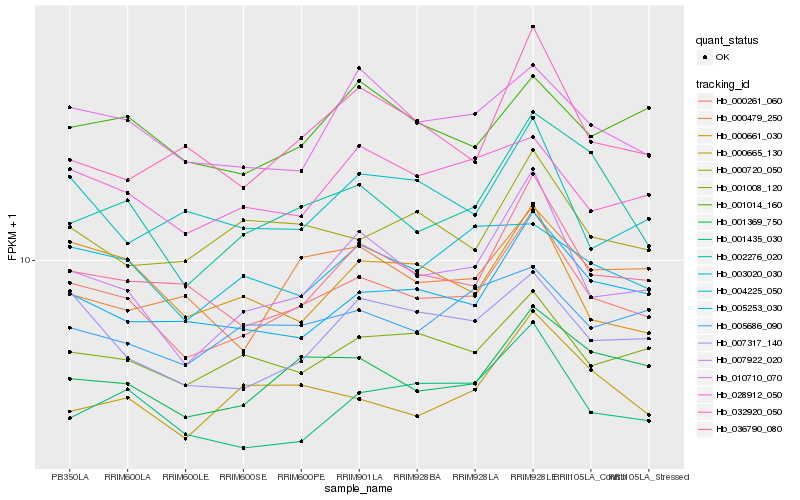
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_060 |
0.0 |
- |
- |
PREDICTED: pre-mRNA-splicing factor 38B-like [Jatropha curcas] |
| 2 |
Hb_007922_020 |
0.0601608414 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 3 |
Hb_036790_080 |
0.069372368 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 4 |
Hb_001369_750 |
0.0711771564 |
- |
- |
PREDICTED: probable rRNA-processing protein EBP2 homolog [Jatropha curcas] |
| 5 |
Hb_005253_030 |
0.0734468668 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 6 |
Hb_001008_120 |
0.075889146 |
desease resistance |
Gene Name: PEX-1N |
peroxisome biogenesis factor, putative [Ricinus communis] |
| 7 |
Hb_007317_140 |
0.0812059906 |
- |
- |
expressed protein, putative [Ricinus communis] |
| 8 |
Hb_010710_070 |
0.0812666836 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 9 |
Hb_002276_020 |
0.0829005871 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH49 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001435_030 |
0.0868404588 |
- |
- |
PREDICTED: uncharacterized protein LOC105645073 [Jatropha curcas] |
| 11 |
Hb_028912_050 |
0.0871857906 |
- |
- |
PREDICTED: calmodulin-interacting protein 111 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000661_030 |
0.0902362655 |
- |
- |
PREDICTED: uncharacterized protein LOC105650374 [Jatropha curcas] |
| 13 |
Hb_000720_050 |
0.0913981759 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 14 |
Hb_005686_090 |
0.0931169221 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 2 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001014_160 |
0.0934287626 |
- |
- |
hypothetical protein JCGZ_23092 [Jatropha curcas] |
| 16 |
Hb_004225_050 |
0.0936447765 |
transcription factor |
TF Family: MYB-related |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_003020_030 |
0.0937927122 |
- |
- |
PREDICTED: protein TOC75-3, chloroplastic-like [Populus euphratica] |
| 18 |
Hb_000479_250 |
0.0938479142 |
- |
- |
PREDICTED: uncharacterized protein LOC105644121 [Jatropha curcas] |
| 19 |
Hb_000665_130 |
0.0939861014 |
- |
- |
PREDICTED: uncharacterized protein LOC105637599 isoform X2 [Jatropha curcas] |
| 20 |
Hb_032920_050 |
0.0956295752 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |