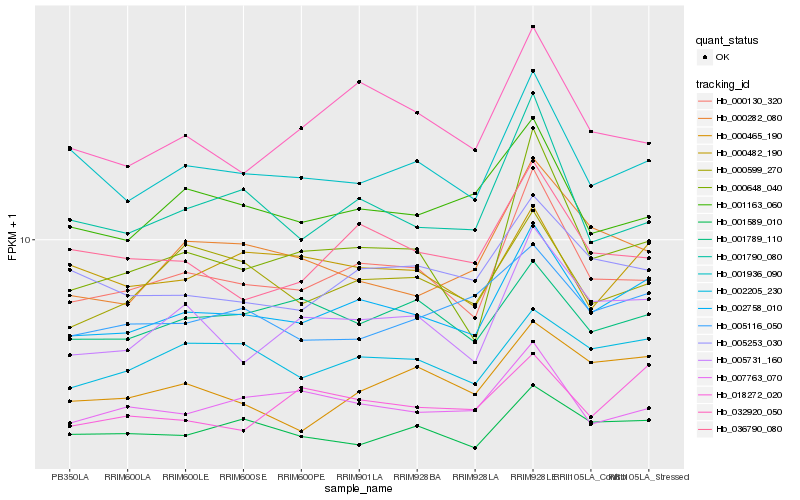
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002758_010 |
0.0 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 2 |
Hb_000130_320 |
0.0668992322 |
desease resistance |
Gene Name: AAA |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 3 |
Hb_002205_230 |
0.0771490792 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_005731_160 |
0.078509611 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 5 |
Hb_001936_090 |
0.0787370085 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 6 |
Hb_005253_030 |
0.0848790288 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_001589_010 |
0.0851892508 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 8 |
Hb_001163_060 |
0.0878379581 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 9 |
Hb_005116_050 |
0.0889751685 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_007763_070 |
0.0899100743 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71460, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000482_190 |
0.091016464 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 12 |
Hb_001789_110 |
0.0910932703 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 13 |
Hb_001790_080 |
0.0911401564 |
- |
- |
PREDICTED: chloroplastic group IIA intron splicing facilitator CRS1, chloroplastic isoform X3 [Jatropha curcas] |
| 14 |
Hb_036790_080 |
0.0913504532 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 39 [Jatropha curcas] |
| 15 |
Hb_000599_270 |
0.0939793119 |
- |
- |
PREDICTED: serine/threonine protein phosphatase 2A regulatory subunit B''beta isoform X3 [Jatropha curcas] |
| 16 |
Hb_032920_050 |
0.0942793304 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 17 |
Hb_018272_020 |
0.0952627562 |
- |
- |
PREDICTED: small glutamine-rich tetratricopeptide repeat-containing protein alpha [Jatropha curcas] |
| 18 |
Hb_000282_080 |
0.0966151532 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000648_040 |
0.0968314117 |
- |
- |
unknown [Populus trichocarpa] |
| 20 |
Hb_000465_190 |
0.0975063921 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HOS1 isoform X2 [Jatropha curcas] |