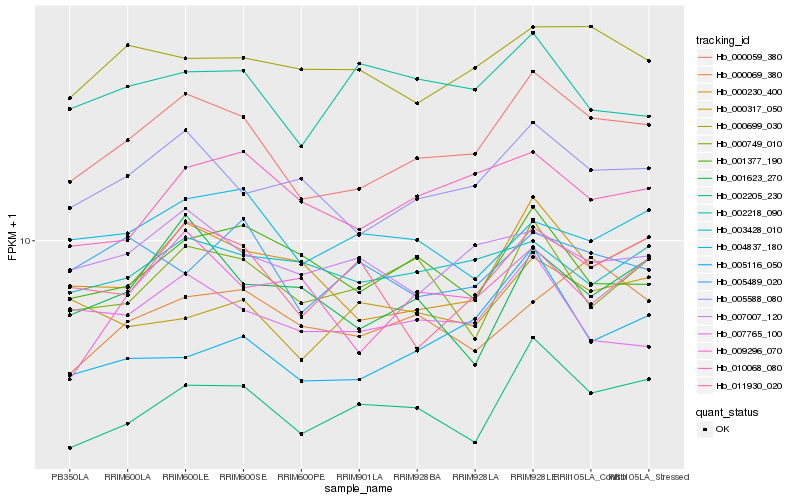
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000059_380 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |
| 2 |
Hb_005588_080 |
0.0614319152 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 3 |
Hb_002205_230 |
0.0664286822 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 4 |
Hb_000230_400 |
0.069228175 |
- |
- |
PREDICTED: protein ABCI12, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_010068_080 |
0.0794796352 |
- |
- |
PREDICTED: RNA-binding protein rsd1-like [Jatropha curcas] |
| 6 |
Hb_007007_120 |
0.0803579901 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000699_030 |
0.081377353 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_011930_020 |
0.0839833427 |
- |
- |
PREDICTED: uncharacterized protein LOC105649787 isoform X1 [Jatropha curcas] |
| 9 |
Hb_007765_100 |
0.0844134867 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 10 |
Hb_000749_010 |
0.0845101563 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_003428_010 |
0.0848044261 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 12 |
Hb_001377_190 |
0.0870800221 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_000317_050 |
0.0872104951 |
- |
- |
formiminotransferase-cyclodeaminase, putative [Ricinus communis] |
| 14 |
Hb_005116_050 |
0.0872248929 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 15 |
Hb_004837_180 |
0.0886394816 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_000069_380 |
0.0887033969 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 17 |
Hb_009296_070 |
0.0902773596 |
- |
- |
PREDICTED: cyclin-dependent kinase D-3 isoform X1 [Jatropha curcas] |
| 18 |
Hb_005489_020 |
0.0912427836 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001623_270 |
0.0912990076 |
transcription factor |
TF Family: PHD |
Inhibitor of growth protein, putative [Ricinus communis] |
| 20 |
Hb_002218_090 |
0.0915194517 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |