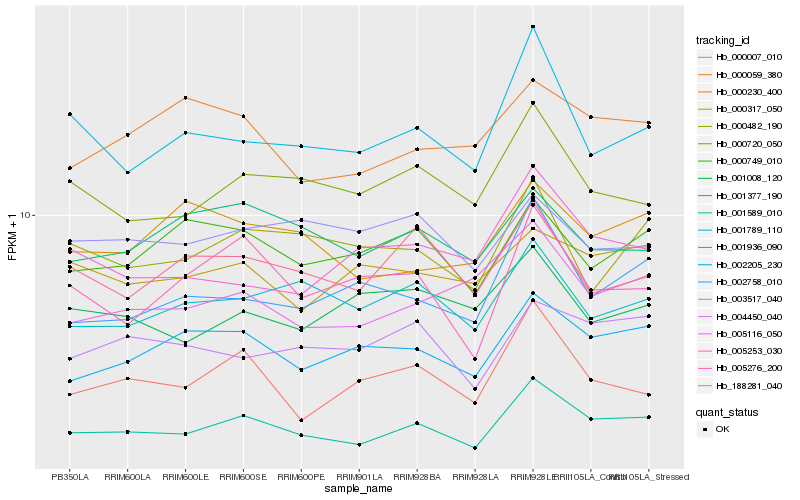
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001589_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638192 isoform X1 [Jatropha curcas] |
| 2 |
Hb_002205_230 |
0.0751375491 |
- |
- |
glycerol-3-phosphate dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_000720_050 |
0.0792882028 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001936_090 |
0.0813009552 |
- |
- |
PREDICTED: BOI-related E3 ubiquitin-protein ligase 1-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_005116_050 |
0.0827620208 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g02820, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_001789_110 |
0.0837549528 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 7 |
Hb_004450_040 |
0.0848011613 |
- |
- |
PREDICTED: DNA ligase 1 [Jatropha curcas] |
| 8 |
Hb_002758_010 |
0.0851892508 |
- |
- |
PREDICTED: ribonuclease II, chloroplastic/mitochondrial [Populus euphratica] |
| 9 |
Hb_000007_010 |
0.0886489952 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000749_010 |
0.0898074006 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 11 |
Hb_188281_040 |
0.0900022572 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 12 |
Hb_000317_050 |
0.0908502833 |
- |
- |
formiminotransferase-cyclodeaminase, putative [Ricinus communis] |
| 13 |
Hb_000482_190 |
0.0912456378 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 14 |
Hb_005276_200 |
0.0914046457 |
- |
- |
PREDICTED: preprotein translocase subunit SCY2, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000230_400 |
0.0922914259 |
- |
- |
PREDICTED: protein ABCI12, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000059_380 |
0.0932289075 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |
| 17 |
Hb_001377_190 |
0.0934957144 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 18 |
Hb_005253_030 |
0.0956998582 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 19 |
Hb_001008_120 |
0.0991885872 |
desease resistance |
Gene Name: PEX-1N |
peroxisome biogenesis factor, putative [Ricinus communis] |
| 20 |
Hb_003517_040 |
0.0995693611 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |