| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
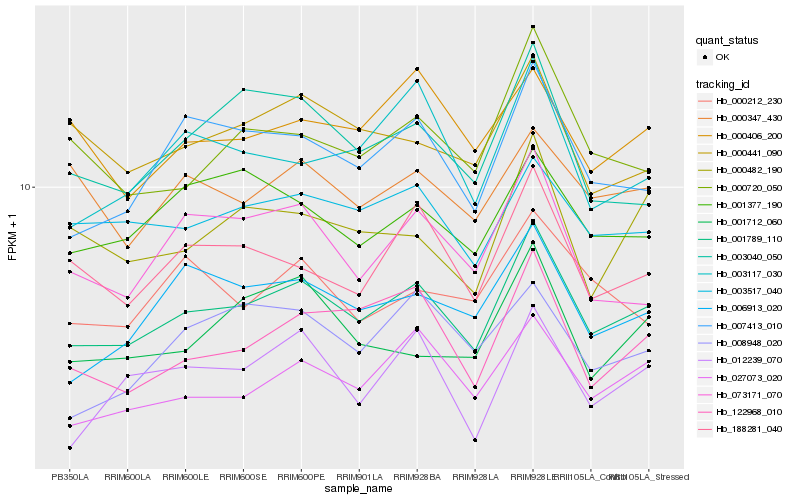
Hb_001789_110 |
0.0 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 2 |
Hb_027073_020 |
0.0521859531 |
- |
- |
PREDICTED: testis-expressed sequence 10 protein isoform X2 [Jatropha curcas] |
| 3 |
Hb_003517_040 |
0.0573454536 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 4 |
Hb_007413_010 |
0.0575443053 |
- |
- |
PREDICTED: SUMO-activating enzyme subunit 2 [Jatropha curcas] |
| 5 |
Hb_122968_010 |
0.0600580067 |
- |
- |
hypothetical protein JCGZ_23576 [Jatropha curcas] |
| 6 |
Hb_188281_040 |
0.0609719363 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 7 |
Hb_006913_020 |
0.0620687042 |
- |
- |
PREDICTED: uncharacterized protein LOC105649145 isoform X1 [Jatropha curcas] |
| 8 |
Hb_000720_050 |
0.0627569997 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001377_190 |
0.0647629057 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 10 |
Hb_003117_030 |
0.067253523 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 11 |
Hb_000482_190 |
0.0681515888 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 [Jatropha curcas] |
| 12 |
Hb_000441_090 |
0.0683211924 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 13 |
Hb_003040_050 |
0.0684601946 |
- |
- |
PREDICTED: uncharacterized protein LOC105638219 [Jatropha curcas] |
| 14 |
Hb_001712_060 |
0.0686598519 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK9 [Jatropha curcas] |
| 15 |
Hb_012239_070 |
0.0694207619 |
- |
- |
PREDICTED: uncharacterized protein LOC105634113 [Jatropha curcas] |
| 16 |
Hb_000347_430 |
0.0697673436 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 17 |
Hb_073171_070 |
0.0697986131 |
- |
- |
PREDICTED: RNA polymerase II-associated factor 1 homolog [Jatropha curcas] |
| 18 |
Hb_000406_200 |
0.0701181119 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 19 |
Hb_000212_230 |
0.0715668155 |
- |
- |
PREDICTED: endoribonuclease Dicer homolog 2 [Jatropha curcas] |
| 20 |
Hb_008948_020 |
0.0717096957 |
- |
- |
hypothetical protein JCGZ_21216 [Jatropha curcas] |