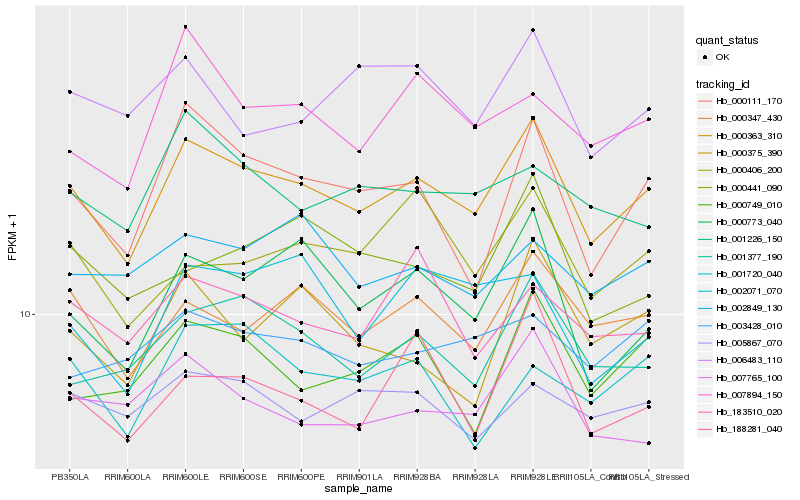
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000363_310 |
0.0 |
- |
- |
DNA topoisomerase type I, putative [Ricinus communis] |
| 2 |
Hb_000347_430 |
0.0556367306 |
- |
- |
PREDICTED: mitochondrial ubiquitin ligase activator of nfkb 1-A [Jatropha curcas] |
| 3 |
Hb_001377_190 |
0.0600939189 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000773_040 |
0.0608114348 |
- |
- |
PREDICTED: uncharacterized protein LOC105641863 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000111_170 |
0.0627577296 |
- |
- |
PREDICTED: uncharacterized protein LOC105631234 isoform X1 [Jatropha curcas] |
| 6 |
Hb_007765_100 |
0.0651042484 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 7 |
Hb_000749_010 |
0.0653339939 |
- |
- |
PREDICTED: non-canonical poly(A) RNA polymerase PAPD5 isoform X1 [Jatropha curcas] |
| 8 |
Hb_007894_150 |
0.0663675906 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 9 |
Hb_188281_040 |
0.0666905009 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 10 |
Hb_003428_010 |
0.0683650807 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 11 |
Hb_001226_150 |
0.0686859394 |
- |
- |
PREDICTED: serine/threonine-protein kinase 38-like isoform X3 [Jatropha curcas] |
| 12 |
Hb_005867_070 |
0.0700205009 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 13 |
Hb_183510_020 |
0.0700417141 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 14 |
Hb_000406_200 |
0.0703262501 |
- |
- |
PREDICTED: translation initiation factor eIF-2B subunit epsilon-like isoform X4 [Jatropha curcas] |
| 15 |
Hb_002849_130 |
0.0707525346 |
- |
- |
PREDICTED: RAB6A-GEF complex partner protein 1-like [Jatropha curcas] |
| 16 |
Hb_001720_040 |
0.0709956688 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000441_090 |
0.0714122254 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 18 |
Hb_000375_390 |
0.0721491681 |
- |
- |
poly-A binding protein, putative [Ricinus communis] |
| 19 |
Hb_006483_110 |
0.0722419557 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 20 |
Hb_002071_070 |
0.0727858187 |
transcription factor |
TF Family: RB |
conserved hypothetical protein [Ricinus communis] |