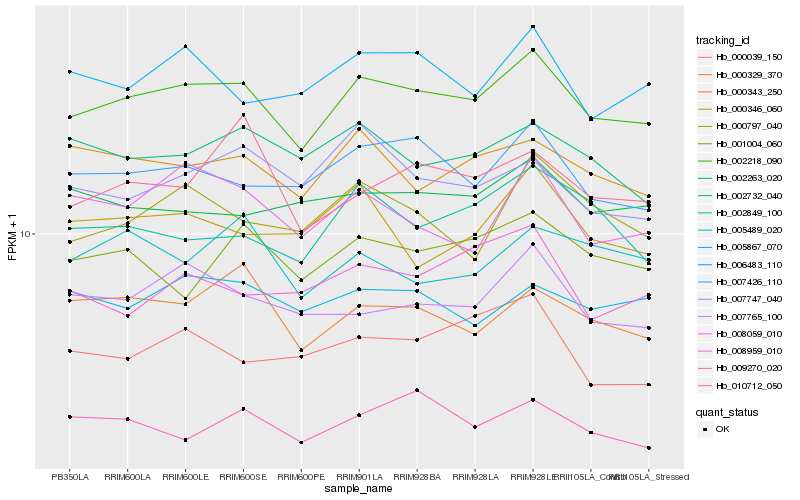
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002218_090 |
0.0 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 2 |
Hb_007426_110 |
0.0573707381 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 3 |
Hb_000343_250 |
0.0635731935 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 4 |
Hb_007765_100 |
0.0649406123 |
- |
- |
PREDICTED: helicase SKI2W [Jatropha curcas] |
| 5 |
Hb_000329_370 |
0.0658255826 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 6 |
Hb_006483_110 |
0.0673398167 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 7 |
Hb_005867_070 |
0.0681084216 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 8 |
Hb_002732_040 |
0.0683298054 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001004_060 |
0.0689865753 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 10 |
Hb_000797_040 |
0.0689902798 |
- |
- |
PREDICTED: uncharacterized protein LOC105645914 [Jatropha curcas] |
| 11 |
Hb_002849_100 |
0.0702965665 |
- |
- |
PREDICTED: mechanosensitive ion channel protein 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_002263_020 |
0.0704083051 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 13 |
Hb_009270_020 |
0.0707760726 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 14 |
Hb_008059_010 |
0.0711824603 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g04810, chloroplastic [Jatropha curcas] |
| 15 |
Hb_007747_040 |
0.0737883272 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 16 |
Hb_010712_050 |
0.0750244616 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 17 |
Hb_000346_060 |
0.0754850006 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 18 |
Hb_000039_150 |
0.0754858341 |
- |
- |
PREDICTED: lysine-specific demethylase JMJ25 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005489_020 |
0.0764139892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_008959_010 |
0.0764234371 |
- |
- |
conserved hypothetical protein [Ricinus communis] |