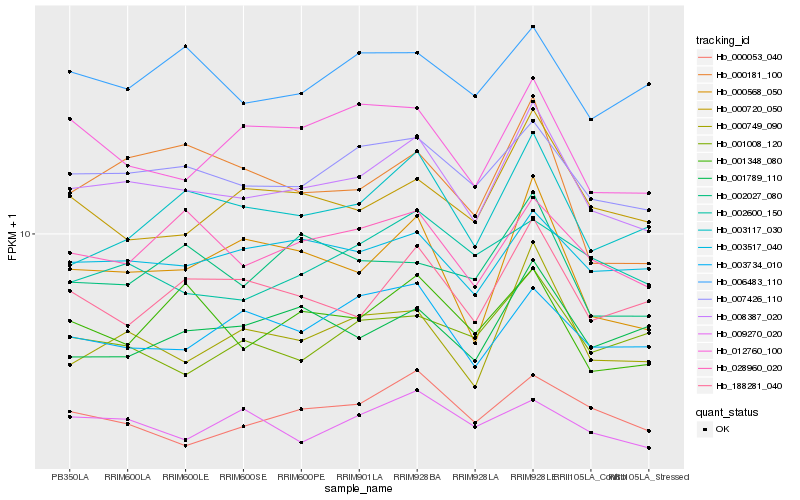
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008387_020 |
0.0 |
- |
- |
PREDICTED: FHA domain-containing protein DDL isoform X1 [Jatropha curcas] |
| 2 |
Hb_007426_110 |
0.0536105353 |
- |
- |
PREDICTED: vacuolar fusion protein CCZ1 homolog isoform X2 [Jatropha curcas] |
| 3 |
Hb_000749_090 |
0.0642898 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 4 |
Hb_003517_040 |
0.0669908919 |
- |
- |
PREDICTED: putative DUF21 domain-containing protein At3g13070, chloroplastic [Jatropha curcas] |
| 5 |
Hb_028960_020 |
0.0682073264 |
- |
- |
PREDICTED: uncharacterized protein LOC105646163 isoform X1 [Jatropha curcas] |
| 6 |
Hb_188281_040 |
0.0696204379 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 12 [Jatropha curcas] |
| 7 |
Hb_000568_050 |
0.0711596069 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_003117_030 |
0.0716950465 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 9 |
Hb_000720_050 |
0.0735350607 |
- |
- |
PREDICTED: uncharacterized protein LOC105645857 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000053_040 |
0.0740799607 |
- |
- |
hypothetical protein CISIN_1g0010392mg, partial [Citrus sinensis] |
| 11 |
Hb_012760_100 |
0.0755510294 |
- |
- |
Pre-rRNA-processing protein ESF1, putative [Ricinus communis] |
| 12 |
Hb_003734_010 |
0.0756237715 |
- |
- |
PREDICTED: double-strand break repair protein MRE11 [Jatropha curcas] |
| 13 |
Hb_001348_080 |
0.0784991708 |
- |
- |
PREDICTED: sister chromatid cohesion protein PDS5 homolog A isoform X1 [Jatropha curcas] |
| 14 |
Hb_001789_110 |
0.0789534522 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_006483_110 |
0.0799188762 |
- |
- |
PREDICTED: RNA-binding protein Musashi homolog 1 [Jatropha curcas] |
| 16 |
Hb_001008_120 |
0.0801625761 |
desease resistance |
Gene Name: PEX-1N |
peroxisome biogenesis factor, putative [Ricinus communis] |
| 17 |
Hb_002027_080 |
0.0801647112 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105650047 [Jatropha curcas] |
| 18 |
Hb_000181_100 |
0.0822633799 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 19 |
Hb_009270_020 |
0.0826333244 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: helicase and polymerase-containing protein TEBICHI [Jatropha curcas] |
| 20 |
Hb_002600_150 |
0.0833148135 |
- |
- |
PREDICTED: uncharacterized protein LOC105646805 isoform X1 [Jatropha curcas] |