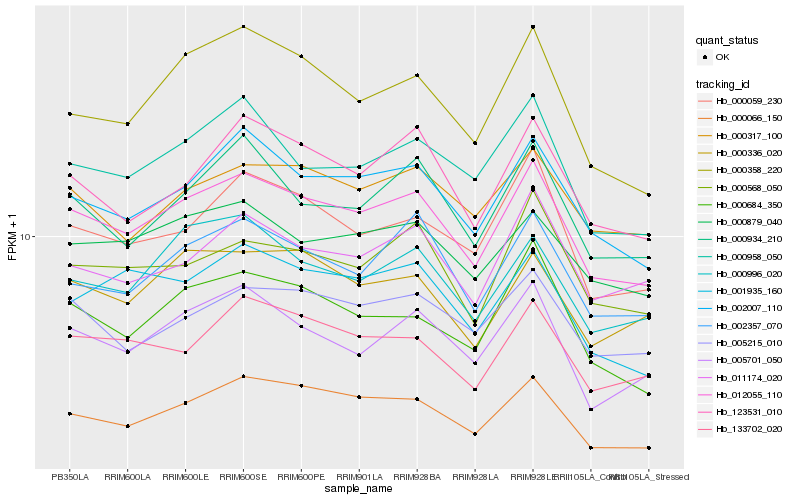
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012055_110 |
0.0 |
- |
- |
tetratricopeptide repeat protein, tpr, putative [Ricinus communis] |
| 2 |
Hb_002007_110 |
0.0478311855 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000066_150 |
0.0500914925 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein POPTR_0004s20690g [Populus trichocarpa] |
| 4 |
Hb_000879_040 |
0.0598880869 |
- |
- |
PREDICTED: calcium-transporting ATPase 3, endoplasmic reticulum-type [Jatropha curcas] |
| 5 |
Hb_000958_050 |
0.060537035 |
- |
- |
hypothetical protein JCGZ_16781 [Jatropha curcas] |
| 6 |
Hb_002357_070 |
0.0618815261 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 7 |
Hb_005701_050 |
0.062049878 |
- |
- |
PREDICTED: protein LTV1 homolog [Jatropha curcas] |
| 8 |
Hb_005215_010 |
0.0630400165 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000934_210 |
0.0640902791 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 10 |
Hb_000358_220 |
0.064116496 |
- |
- |
PREDICTED: SNW/SKI-interacting protein-like [Jatropha curcas] |
| 11 |
Hb_123531_010 |
0.0642312576 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 12 |
Hb_000059_230 |
0.0642680517 |
- |
- |
PREDICTED: uncharacterized protein LOC105648049 [Jatropha curcas] |
| 13 |
Hb_133702_020 |
0.0647677483 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 14 |
Hb_000996_020 |
0.0659602231 |
- |
- |
PREDICTED: RNA-binding protein NOB1 [Jatropha curcas] |
| 15 |
Hb_000684_350 |
0.0663135278 |
- |
- |
PREDICTED: uncharacterized protein LOC105642163 isoform X1 [Jatropha curcas] |
| 16 |
Hb_000568_050 |
0.0667851602 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000317_100 |
0.0672026955 |
- |
- |
PREDICTED: uncharacterized protein LOC105646995 [Jatropha curcas] |
| 18 |
Hb_011174_020 |
0.0679992383 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 19 |
Hb_001935_160 |
0.0681093356 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 20 |
Hb_000336_020 |
0.0681295876 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |