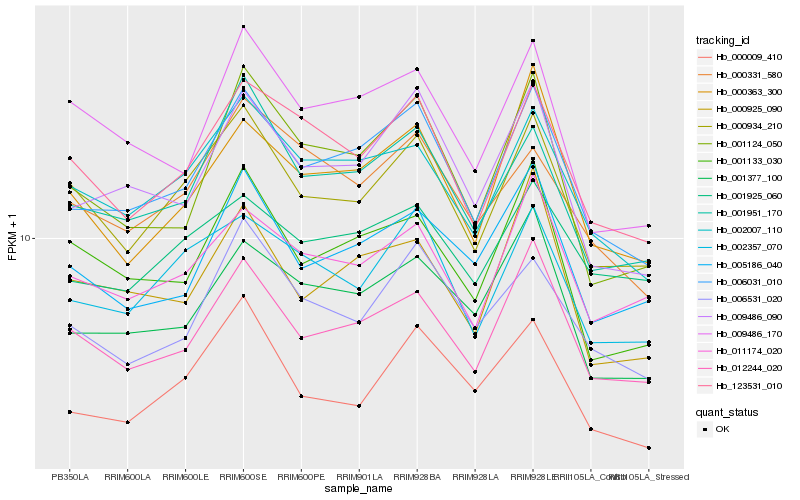
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006031_010 |
0.0 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 2 |
Hb_009486_090 |
0.0453480497 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 3 |
Hb_002007_110 |
0.0555493516 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25 isoform X2 [Jatropha curcas] |
| 4 |
Hb_012244_020 |
0.0558481794 |
- |
- |
calpain, putative [Ricinus communis] |
| 5 |
Hb_001377_100 |
0.0561331211 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 6 |
Hb_001951_170 |
0.0563394493 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 7 |
Hb_123531_010 |
0.0602699453 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 8 |
Hb_000934_210 |
0.0647637613 |
- |
- |
suppressor of ty, putative [Ricinus communis] |
| 9 |
Hb_000363_300 |
0.0650218462 |
- |
- |
PREDICTED: protein ENHANCED DOWNY MILDEW 2 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001124_050 |
0.0661574827 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 11 |
Hb_002357_070 |
0.0669402206 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 12 |
Hb_006531_020 |
0.0674882585 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 13 |
Hb_005186_040 |
0.0678866088 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 14 |
Hb_011174_020 |
0.0697761546 |
- |
- |
PREDICTED: uncharacterized protein LOC105632610 [Jatropha curcas] |
| 15 |
Hb_001133_030 |
0.0717299239 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 16 |
Hb_000925_090 |
0.0720084712 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000009_410 |
0.0721756676 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 18 |
Hb_009486_170 |
0.0735467016 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 19 |
Hb_000331_580 |
0.0739299086 |
- |
- |
PREDICTED: programmed cell death protein 4-like [Jatropha curcas] |
| 20 |
Hb_001925_060 |
0.0742268784 |
- |
- |
PREDICTED: putative RNA polymerase II subunit B1 CTD phosphatase RPAP2 homolog [Jatropha curcas] |