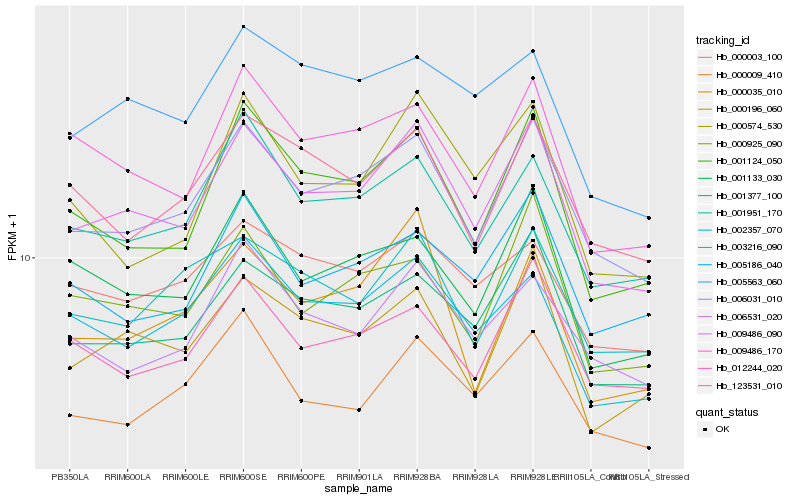
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009486_090 |
0.0 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_006031_010 |
0.0453480497 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15b isoform X3 [Jatropha curcas] |
| 3 |
Hb_001377_100 |
0.0549795896 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001124_050 |
0.0605415903 |
transcription factor |
TF Family: SNF2 |
PREDICTED: chromatin structure-remodeling complex protein SYD-like [Jatropha curcas] |
| 5 |
Hb_000196_060 |
0.061512845 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 6 |
Hb_001951_170 |
0.0689923806 |
- |
- |
PREDICTED: uncharacterized protein LOC105649755 [Jatropha curcas] |
| 7 |
Hb_009486_170 |
0.0705055466 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 4G [Jatropha curcas] |
| 8 |
Hb_001133_030 |
0.0726359713 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 9 |
Hb_003216_090 |
0.0742071088 |
- |
- |
PREDICTED: nuclear pore complex protein NUP133 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000925_090 |
0.0760972125 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 11 |
Hb_012244_020 |
0.0768487994 |
- |
- |
calpain, putative [Ricinus communis] |
| 12 |
Hb_000009_410 |
0.0776023263 |
transcription factor |
TF Family: SET |
hypothetical protein POPTR_0007s12130g [Populus trichocarpa] |
| 13 |
Hb_005186_040 |
0.0779148334 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 13 isoform X2 [Jatropha curcas] |
| 14 |
Hb_006531_020 |
0.0779349474 |
- |
- |
PREDICTED: autophagy-related protein 13 [Jatropha curcas] |
| 15 |
Hb_000574_530 |
0.0792053702 |
- |
- |
PREDICTED: pre-mRNA-processing-splicing factor 8 [Jatropha curcas] |
| 16 |
Hb_123531_010 |
0.0793547965 |
- |
- |
PREDICTED: nuclear pore complex protein NUP98A isoform X1 [Jatropha curcas] |
| 17 |
Hb_002357_070 |
0.0802705587 |
transcription factor |
TF Family: PHD |
PREDICTED: protein strawberry notch homolog 1 [Jatropha curcas] |
| 18 |
Hb_005563_060 |
0.0805438507 |
- |
- |
PREDICTED: coatomer subunit alpha-1 [Jatropha curcas] |
| 19 |
Hb_000035_010 |
0.0807056604 |
transcription factor |
TF Family: ARR-B |
PREDICTED: two-component response regulator ARR12-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_000003_100 |
0.0812098585 |
transcription factor |
TF Family: SNF2 |
PREDICTED: transcription termination factor 2 isoform X5 [Jatropha curcas] |