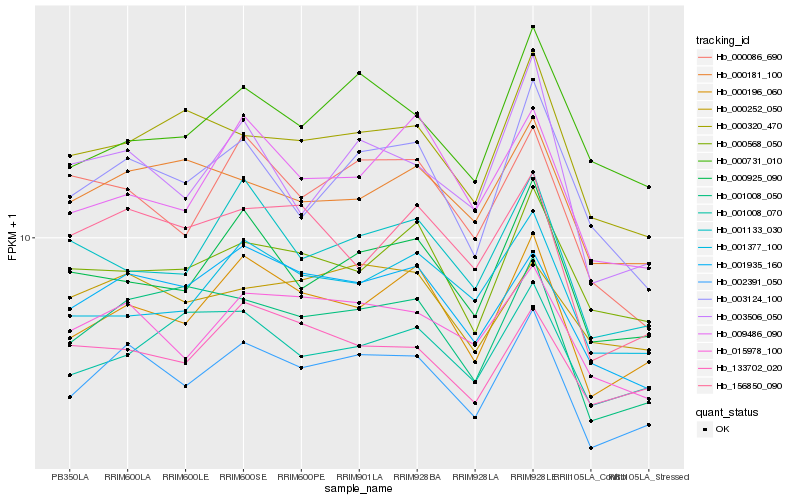
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002391_050 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17670 [Jatropha curcas] |
| 2 |
Hb_000196_060 |
0.0668093448 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |
| 3 |
Hb_001008_050 |
0.0788300567 |
- |
- |
PREDICTED: WD repeat-containing protein 43 [Jatropha curcas] |
| 4 |
Hb_015978_100 |
0.080039641 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH6 [Jatropha curcas] |
| 5 |
Hb_001935_160 |
0.0813179595 |
- |
- |
PREDICTED: F-box protein At5g51370-like [Jatropha curcas] |
| 6 |
Hb_000925_090 |
0.0909237878 |
- |
- |
PREDICTED: uncharacterized protein LOC105635805 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003124_100 |
0.0924047364 |
- |
- |
- |
| 8 |
Hb_003506_050 |
0.0973796797 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_156850_090 |
0.0976770092 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 10 |
Hb_000320_470 |
0.0987031597 |
- |
- |
calcium-dependent protein kinase [Hevea brasiliensis] |
| 11 |
Hb_133702_020 |
0.0993323757 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000181_100 |
0.0996254344 |
- |
- |
PREDICTED: protein starmaker [Jatropha curcas] |
| 13 |
Hb_000252_050 |
0.0999468857 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 14 |
Hb_000086_690 |
0.1000267452 |
- |
- |
PREDICTED: splicing factor 3B subunit 1 [Jatropha curcas] |
| 15 |
Hb_000568_050 |
0.1009060146 |
- |
- |
PREDICTED: endoplasmic reticulum metallopeptidase 1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001133_030 |
0.1033007146 |
- |
- |
PREDICTED: MAG2-interacting protein 2-like [Jatropha curcas] |
| 17 |
Hb_000731_010 |
0.1053695048 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 1, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001008_070 |
0.1053857116 |
- |
- |
PREDICTED: periodic tryptophan protein 2 homolog [Jatropha curcas] |
| 19 |
Hb_001377_100 |
0.1054532602 |
- |
- |
PREDICTED: uncharacterized protein LOC105642863 isoform X1 [Jatropha curcas] |
| 20 |
Hb_009486_090 |
0.1059021711 |
- |
- |
RNA binding protein, putative [Ricinus communis] |