| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
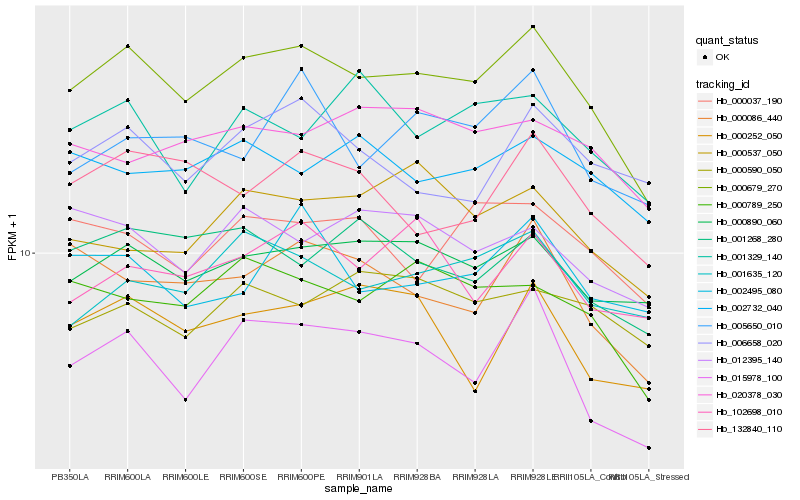
Hb_000679_270 |
0.0 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 2 |
Hb_000890_060 |
0.0715614483 |
- |
- |
PREDICTED: sucrose nonfermenting 4-like protein [Jatropha curcas] |
| 3 |
Hb_015978_100 |
0.0727183442 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH6 [Jatropha curcas] |
| 4 |
Hb_000789_250 |
0.0740845961 |
- |
- |
PREDICTED: uncharacterized protein LOC105637809 [Jatropha curcas] |
| 5 |
Hb_000086_440 |
0.0780282996 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 6 |
Hb_132840_110 |
0.0820060413 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_001635_120 |
0.0835610505 |
- |
- |
PREDICTED: uncharacterized protein At1g51745-like [Jatropha curcas] |
| 8 |
Hb_001268_280 |
0.084431244 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000037_190 |
0.085642557 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 10 |
Hb_102698_010 |
0.0873928705 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 5-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_012395_140 |
0.0876671349 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 12 |
Hb_002732_040 |
0.0882141719 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005650_010 |
0.0886023725 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_020378_030 |
0.0896903372 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 15 |
Hb_006658_020 |
0.0904601318 |
- |
- |
PREDICTED: uncharacterized protein LOC105633529 [Jatropha curcas] |
| 16 |
Hb_002495_080 |
0.0904646953 |
- |
- |
PREDICTED: polygalacturonase ADPG2 [Jatropha curcas] |
| 17 |
Hb_001329_140 |
0.0907529046 |
- |
- |
PREDICTED: zinc finger protein 598 [Jatropha curcas] |
| 18 |
Hb_000252_050 |
0.0917759272 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g60770 [Jatropha curcas] |
| 19 |
Hb_000537_050 |
0.0924277585 |
- |
- |
AP-2 complex subunit beta-1, putative [Ricinus communis] |
| 20 |
Hb_000590_050 |
0.0925702328 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |