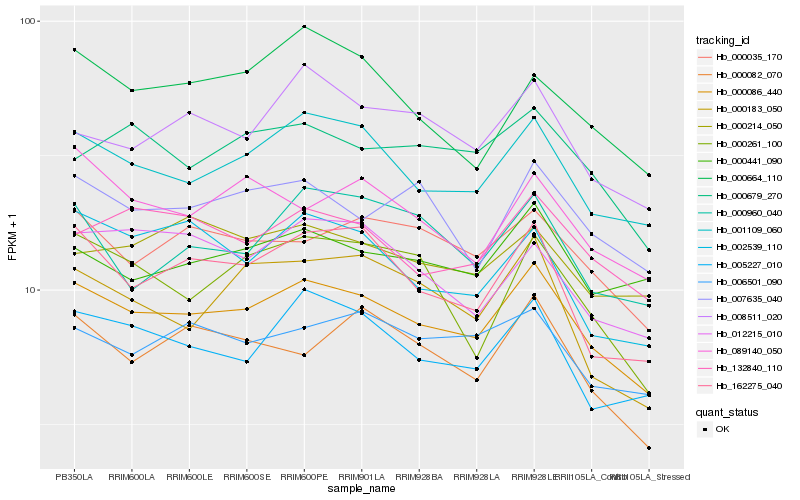
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000086_440 |
0.0 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 2 |
Hb_001109_060 |
0.0495342373 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 3 |
Hb_005227_010 |
0.0676815266 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 4 |
Hb_162275_040 |
0.0689358484 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000035_170 |
0.0707624739 |
- |
- |
PREDICTED: DNA-directed RNA polymerases IV and V subunit 2-like [Jatropha curcas] |
| 6 |
Hb_132840_110 |
0.0718747226 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_006501_090 |
0.0742553591 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 8 |
Hb_007635_040 |
0.0744437737 |
- |
- |
PREDICTED: WAT1-related protein At3g45870 isoform X1 [Jatropha curcas] |
| 9 |
Hb_008511_020 |
0.0761092625 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 10 |
Hb_000441_090 |
0.0775508816 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 11 |
Hb_000679_270 |
0.0780282996 |
- |
- |
PREDICTED: putative clathrin assembly protein At2g25430 [Jatropha curcas] |
| 12 |
Hb_000082_070 |
0.081263681 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 1 [Jatropha curcas] |
| 13 |
Hb_002539_110 |
0.0813654372 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 14 |
Hb_089140_050 |
0.0819103706 |
- |
- |
hypothetical protein JCGZ_07485 [Jatropha curcas] |
| 15 |
Hb_012215_010 |
0.0825661863 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 16 |
Hb_000664_110 |
0.0831658351 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 17 |
Hb_000183_050 |
0.0848784949 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 18 |
Hb_000261_100 |
0.0850024001 |
- |
- |
PREDICTED: beta-galactosidase 9 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000214_050 |
0.0850964146 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000960_040 |
0.0854284851 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |