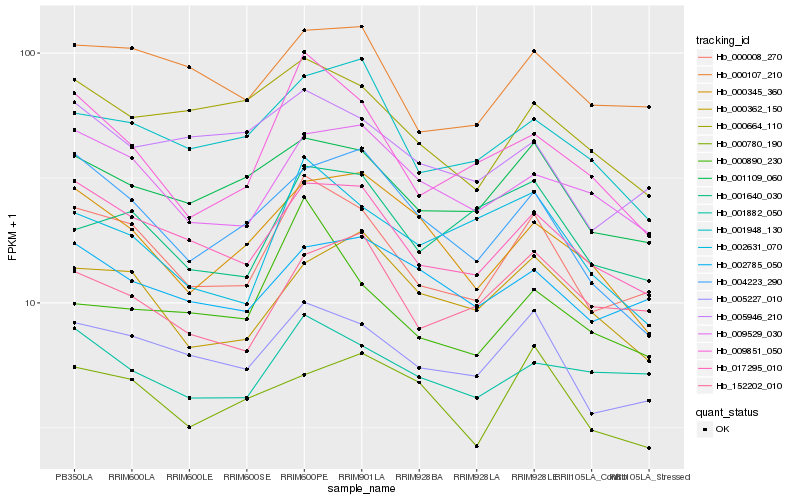
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000008_270 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein REVOLUTA [Jatropha curcas] |
| 2 |
Hb_005227_010 |
0.068780836 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 3 |
Hb_017295_010 |
0.0706561489 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 4 |
Hb_000107_210 |
0.0877744486 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 5 |
Hb_001109_060 |
0.0893827983 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 6 |
Hb_009851_050 |
0.0919142011 |
- |
- |
PREDICTED: nephrocystin-3 [Jatropha curcas] |
| 7 |
Hb_009529_030 |
0.0960970667 |
- |
- |
PREDICTED: transmembrane protein 120 homolog [Jatropha curcas] |
| 8 |
Hb_001882_050 |
0.1017178804 |
- |
- |
hydroxysteroid dehydrogenase, putative [Ricinus communis] |
| 9 |
Hb_001948_130 |
0.1055881002 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 10 |
Hb_002785_050 |
0.1059065659 |
- |
- |
PREDICTED: adenylosuccinate lyase-like [Jatropha curcas] |
| 11 |
Hb_005946_210 |
0.1070615887 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 12 |
Hb_000664_110 |
0.107948541 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 13 |
Hb_152202_010 |
0.1085912643 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 1 [Jatropha curcas] |
| 14 |
Hb_004223_290 |
0.1087861706 |
- |
- |
PREDICTED: protein IQ-DOMAIN 32 [Jatropha curcas] |
| 15 |
Hb_001640_030 |
0.1102805209 |
- |
- |
PREDICTED: uncharacterized protein LOC105630021 [Jatropha curcas] |
| 16 |
Hb_000362_150 |
0.1116692347 |
- |
- |
PREDICTED: homogentisate 1,2-dioxygenase [Jatropha curcas] |
| 17 |
Hb_002631_070 |
0.1128147912 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 18 |
Hb_000345_360 |
0.1141183618 |
- |
- |
PREDICTED: protein AUXIN RESPONSE 4 [Jatropha curcas] |
| 19 |
Hb_000780_190 |
0.1150269877 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |
| 20 |
Hb_000890_230 |
0.1153682901 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |