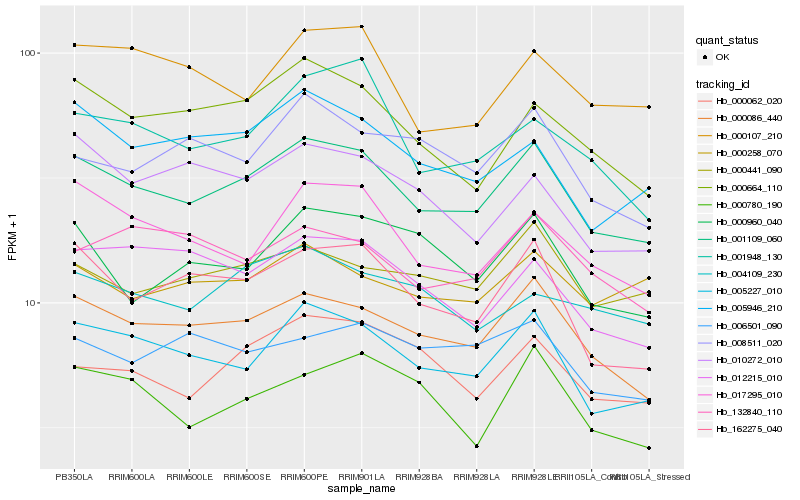
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001109_060 |
0.0 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 2 |
Hb_000086_440 |
0.0495342373 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 3 |
Hb_005227_010 |
0.053102275 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 4 |
Hb_162275_040 |
0.0692078548 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000664_110 |
0.0729009655 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 6 |
Hb_017295_010 |
0.0753466621 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g06890 [Jatropha curcas] |
| 7 |
Hb_005946_210 |
0.0758017644 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 8 |
Hb_000441_090 |
0.0785316476 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 6 regulatory subunit 2 [Jatropha curcas] |
| 9 |
Hb_000062_020 |
0.0798803051 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 10 |
Hb_001948_130 |
0.0831344322 |
- |
- |
PREDICTED: uncharacterized protein LOC105638579 [Jatropha curcas] |
| 11 |
Hb_000960_040 |
0.0835835883 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 12 |
Hb_000107_210 |
0.0837901616 |
- |
- |
PREDICTED: ankyrin repeat domain-containing protein 2 [Jatropha curcas] |
| 13 |
Hb_012215_010 |
0.0854183608 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 14 |
Hb_008511_020 |
0.085606344 |
- |
- |
PREDICTED: coatomer subunit gamma-2 [Jatropha curcas] |
| 15 |
Hb_010272_010 |
0.0858917044 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 16 |
Hb_004109_230 |
0.0864636502 |
- |
- |
heat shock protein binding protein, putative [Ricinus communis] |
| 17 |
Hb_132840_110 |
0.0868490974 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
pyruvate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_006501_090 |
0.0870340419 |
- |
- |
PREDICTED: golgin candidate 1 [Jatropha curcas] |
| 19 |
Hb_000258_070 |
0.0882059292 |
- |
- |
PREDICTED: uncharacterized protein LOC105649104 [Jatropha curcas] |
| 20 |
Hb_000780_190 |
0.0884925424 |
transcription factor |
TF Family: Orphans |
PREDICTED: histidine kinase 2 [Jatropha curcas] |