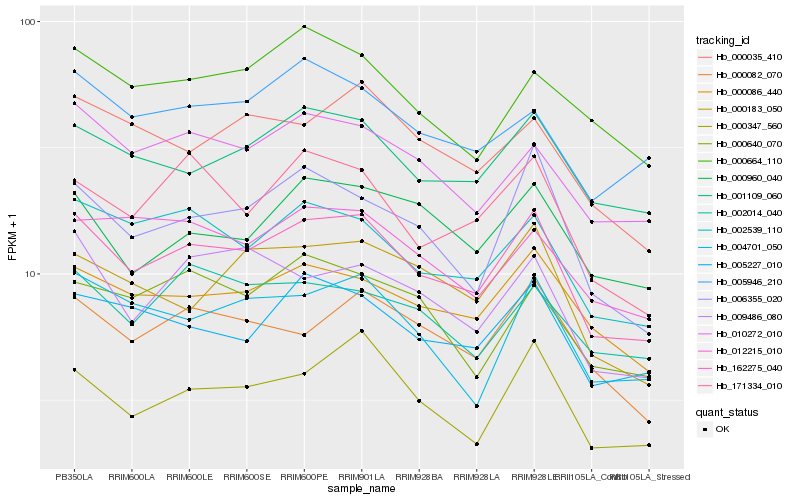
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_162275_040 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000086_440 |
0.0689358484 |
- |
- |
PREDICTED: protein BREAST CANCER SUSCEPTIBILITY 1 homolog [Jatropha curcas] |
| 3 |
Hb_002539_110 |
0.0689405638 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 4 |
Hb_001109_060 |
0.0692078548 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 5 |
Hb_010272_010 |
0.0721172671 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 6 |
Hb_005227_010 |
0.072328226 |
- |
- |
PREDICTED: uncharacterized protein LOC105633453 isoform X1 [Jatropha curcas] |
| 7 |
Hb_006355_020 |
0.0747188879 |
- |
- |
PREDICTED: CLIP-associated protein [Jatropha curcas] |
| 8 |
Hb_000347_560 |
0.0781470526 |
- |
- |
- |
| 9 |
Hb_171334_010 |
0.0795067903 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 10 |
Hb_000082_070 |
0.0796093579 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase alpha 1 [Jatropha curcas] |
| 11 |
Hb_004701_050 |
0.0802697955 |
- |
- |
alpha1 family protein [Populus trichocarpa] |
| 12 |
Hb_000035_410 |
0.0828322531 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit B-like [Jatropha curcas] |
| 13 |
Hb_000960_040 |
0.0837776977 |
- |
- |
PREDICTED: ERAD-associated E3 ubiquitin-protein ligase HRD1B-like [Vitis vinifera] |
| 14 |
Hb_005946_210 |
0.0838420523 |
- |
- |
hypothetical protein JCGZ_07228 [Jatropha curcas] |
| 15 |
Hb_009486_080 |
0.084705403 |
- |
- |
PREDICTED: uncharacterized protein LOC105643242 [Jatropha curcas] |
| 16 |
Hb_000183_050 |
0.0848637906 |
- |
- |
PREDICTED: alpha-mannosidase 2 [Jatropha curcas] |
| 17 |
Hb_012215_010 |
0.0852179116 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 18 |
Hb_000664_110 |
0.0874731415 |
- |
- |
PREDICTED: monoglyceride lipase [Jatropha curcas] |
| 19 |
Hb_000640_070 |
0.0878070795 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 20 |
Hb_002014_040 |
0.0884522438 |
- |
- |
site-1 protease, putative [Ricinus communis] |