| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
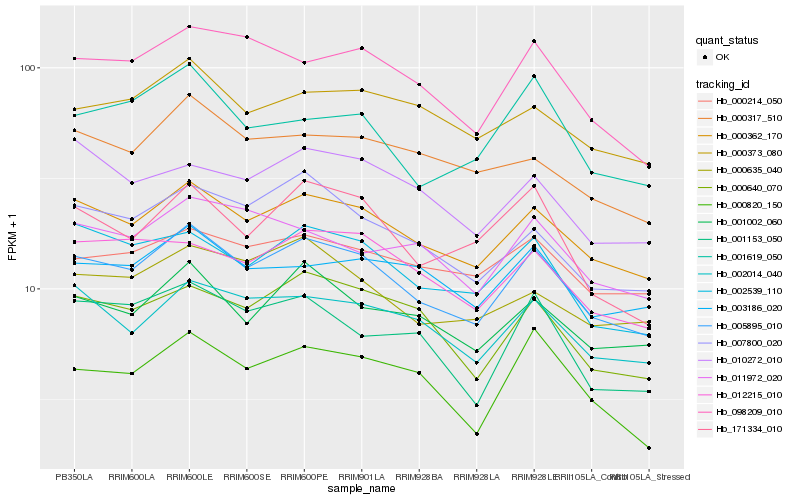
Hb_005895_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000362_170 |
0.0355794329 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 3 |
Hb_002539_110 |
0.0641104836 |
- |
- |
hypothetical protein JCGZ_15697 [Jatropha curcas] |
| 4 |
Hb_007800_020 |
0.065820021 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002014_040 |
0.0663479183 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 6 |
Hb_001002_060 |
0.0669410728 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 7 |
Hb_000640_070 |
0.0682333307 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 8 |
Hb_098209_010 |
0.0703531727 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 9 |
Hb_012215_010 |
0.0705823994 |
- |
- |
PREDICTED: YTH domain-containing family protein 1 [Populus euphratica] |
| 10 |
Hb_171334_010 |
0.07632349 |
- |
- |
PREDICTED: CDPK-related kinase 5 [Jatropha curcas] |
| 11 |
Hb_003186_020 |
0.076934196 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000635_040 |
0.0770969931 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 13 |
Hb_000373_080 |
0.0779507657 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 14 |
Hb_000214_050 |
0.0790822193 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000317_510 |
0.0798040497 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 16 |
Hb_000820_150 |
0.0805561231 |
- |
- |
PREDICTED: ACT domain-containing protein ACR9-like [Jatropha curcas] |
| 17 |
Hb_011972_020 |
0.0808237265 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_010272_010 |
0.081380677 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 19 |
Hb_001619_050 |
0.0820970522 |
- |
- |
PREDICTED: uncharacterized protein LOC105630599 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001153_050 |
0.0823715894 |
- |
- |
PREDICTED: protein FIZZY-RELATED 2 [Jatropha curcas] |