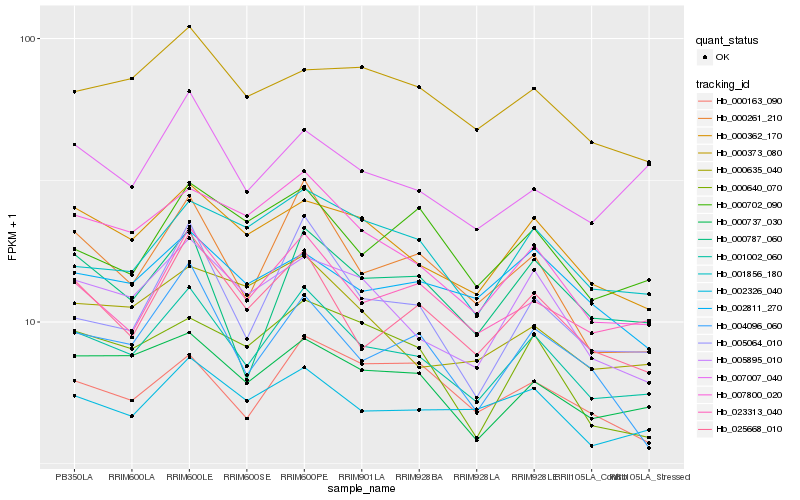
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001002_060 |
0.0 |
- |
- |
PREDICTED: putative GPI-anchor transamidase [Jatropha curcas] |
| 2 |
Hb_023313_040 |
0.0584820156 |
- |
- |
PREDICTED: uncharacterized protein LOC105640827 isoform X2 [Jatropha curcas] |
| 3 |
Hb_000362_170 |
0.0592756471 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 4 |
Hb_000737_030 |
0.0623079125 |
- |
- |
PREDICTED: probable galacturonosyltransferase 3 isoform X1 [Jatropha curcas] |
| 5 |
Hb_025668_010 |
0.0640803849 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000261_210 |
0.0663746619 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 7 |
Hb_005895_010 |
0.0669410728 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002326_040 |
0.0704266039 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os01g0723500-like [Jatropha curcas] |
| 9 |
Hb_007800_020 |
0.0719601482 |
- |
- |
PREDICTED: probable arabinosyltransferase ARAD1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_005064_010 |
0.0728726717 |
- |
- |
PREDICTED: kelch repeat-containing protein At3g27220-like [Jatropha curcas] |
| 11 |
Hb_001856_180 |
0.076082046 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit M [Jatropha curcas] |
| 12 |
Hb_000163_090 |
0.0767560867 |
- |
- |
gamma-tubulin complex component, putative [Ricinus communis] |
| 13 |
Hb_000373_080 |
0.0768053902 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 14 |
Hb_007007_040 |
0.0782676615 |
desease resistance |
Gene Name: ArsA_ATPase |
arsenical pump-driving atpase, putative [Ricinus communis] |
| 15 |
Hb_004096_060 |
0.0787460436 |
- |
- |
PREDICTED: probable dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase isoform X2 [Jatropha curcas] |
| 16 |
Hb_000787_060 |
0.079069091 |
- |
- |
thioredoxin domain-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000702_090 |
0.0800506354 |
- |
- |
26S proteasome non-ATPase regulatory subunit 11 [Theobroma cacao] |
| 18 |
Hb_002811_270 |
0.0803897383 |
- |
- |
PREDICTED: peptide-N(4)-(N-acetyl-beta-glucosaminyl)asparagine amidase [Jatropha curcas] |
| 19 |
Hb_000635_040 |
0.080587419 |
- |
- |
PREDICTED: nuclear pore complex protein NUP54 [Jatropha curcas] |
| 20 |
Hb_000640_070 |
0.0812773316 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |