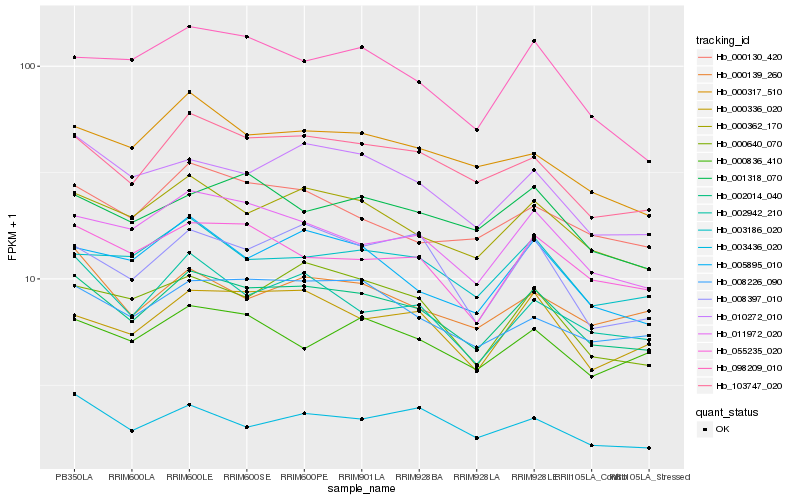
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002014_040 |
0.0 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 2 |
Hb_000362_170 |
0.0481211951 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 3 |
Hb_103747_020 |
0.0516399335 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 4 |
Hb_011972_020 |
0.0600105602 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_010272_010 |
0.060884774 |
- |
- |
PREDICTED: endoplasmic reticulum-Golgi intermediate compartment protein 3 [Jatropha curcas] |
| 6 |
Hb_008226_090 |
0.0649329433 |
- |
- |
PREDICTED: reticulon-like protein B16 isoform X1 [Jatropha curcas] |
| 7 |
Hb_005895_010 |
0.0663479183 |
- |
- |
PREDICTED: uncharacterized protein LOC105643387 isoform X1 [Jatropha curcas] |
| 8 |
Hb_055235_020 |
0.0664879084 |
- |
- |
PREDICTED: probable mitochondrial saccharopine dehydrogenase-like oxidoreductase At5g39410 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002942_210 |
0.0690484311 |
- |
- |
alpha-2,8-sialyltransferase 8b, putative [Ricinus communis] |
| 10 |
Hb_003186_020 |
0.0694332761 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000336_020 |
0.0698459053 |
- |
- |
PREDICTED: phospholipid-transporting ATPase 2 [Jatropha curcas] |
| 12 |
Hb_000317_510 |
0.070786246 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 13 |
Hb_000130_420 |
0.0717568862 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 14 |
Hb_000139_260 |
0.0725021578 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 15 |
Hb_098209_010 |
0.0732533898 |
- |
- |
ATP synthase subunit beta vacuolar, putative [Ricinus communis] |
| 16 |
Hb_000640_070 |
0.0734145127 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 51 homolog [Jatropha curcas] |
| 17 |
Hb_003436_020 |
0.0736106012 |
transcription factor |
TF Family: Orphans |
PREDICTED: uncharacterized protein LOC105636276 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000836_410 |
0.0742620177 |
- |
- |
sec10, putative [Ricinus communis] |
| 19 |
Hb_008397_010 |
0.0745718937 |
- |
- |
PREDICTED: uncharacterized protein LOC105640192 isoform X2 [Jatropha curcas] |
| 20 |
Hb_001318_070 |
0.0753132775 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |