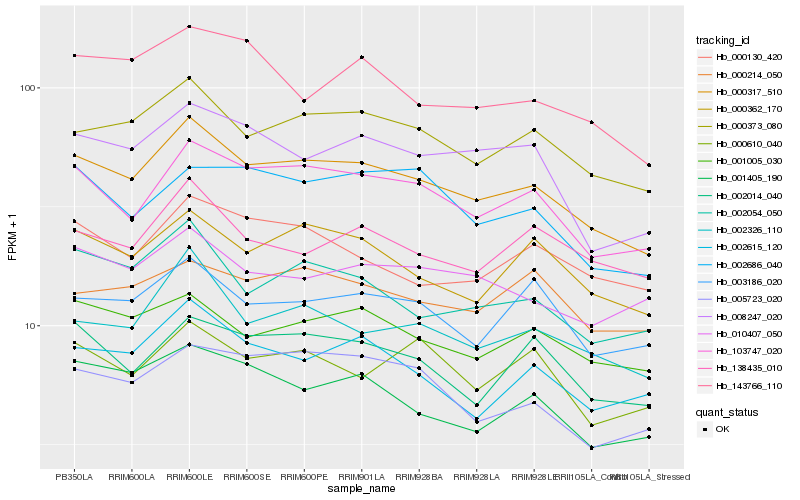
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000317_510 |
0.0 |
- |
- |
PREDICTED: long chain base biosynthesis protein 1 [Jatropha curcas] |
| 2 |
Hb_103747_020 |
0.0493504376 |
- |
- |
calcium dependent protein kinase, partial [Hevea brasiliensis] |
| 3 |
Hb_000373_080 |
0.0498676271 |
- |
- |
PREDICTED: serine decarboxylase [Jatropha curcas] |
| 4 |
Hb_002326_110 |
0.0621136175 |
- |
- |
PREDICTED: uncharacterized protein LOC103320920 [Prunus mume] |
| 5 |
Hb_000362_170 |
0.0629159925 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 6 |
Hb_001005_030 |
0.0648843223 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 7 |
Hb_002054_050 |
0.0672801871 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 8 |
Hb_008247_020 |
0.0692554081 |
- |
- |
PREDICTED: cyclin-L1-1-like [Jatropha curcas] |
| 9 |
Hb_002014_040 |
0.070786246 |
- |
- |
site-1 protease, putative [Ricinus communis] |
| 10 |
Hb_003186_020 |
0.0713917619 |
- |
- |
PREDICTED: histone deacetylase 15 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001405_190 |
0.0719404281 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 12 |
Hb_002615_120 |
0.0721289265 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_005723_020 |
0.0740528895 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_010407_050 |
0.0749040381 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 15 |
Hb_000130_420 |
0.0750022483 |
- |
- |
PREDICTED: uncharacterized protein LOC105635728 [Jatropha curcas] |
| 16 |
Hb_138435_010 |
0.075694466 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
brg-1 associated factor, putative [Ricinus communis] |
| 17 |
Hb_143766_110 |
0.0758088969 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 1 [Jatropha curcas] |
| 18 |
Hb_002686_040 |
0.0765787234 |
- |
- |
PREDICTED: T-complex protein 1 subunit theta [Jatropha curcas] |
| 19 |
Hb_000610_040 |
0.0785174084 |
- |
- |
PREDICTED: uncharacterized protein LOC105635307 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000214_050 |
0.0786367619 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1 isoform X1 [Jatropha curcas] |