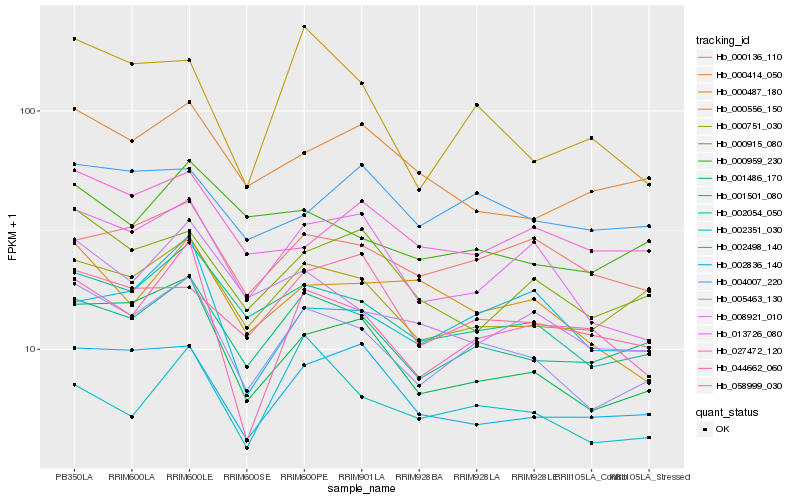
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005463_130 |
0.0 |
- |
- |
PREDICTED: polygalacturonate 4-alpha-galacturonosyltransferase [Jatropha curcas] |
| 2 |
Hb_001486_170 |
0.0696645374 |
- |
- |
PREDICTED: syntaxin-132 [Jatropha curcas] |
| 3 |
Hb_002054_050 |
0.0704543775 |
- |
- |
PREDICTED: uncharacterized protein LOC105641285 [Jatropha curcas] |
| 4 |
Hb_001501_080 |
0.0765149871 |
- |
- |
PREDICTED: uncharacterized protein LOC105640832 isoform X1 [Jatropha curcas] |
| 5 |
Hb_013726_080 |
0.0866608475 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 6 |
Hb_000751_030 |
0.0908627691 |
- |
- |
Vesicle transport protein GOT1B, putative [Ricinus communis] |
| 7 |
Hb_008921_010 |
0.0952570153 |
- |
- |
PREDICTED: uncharacterized protein LOC105629323 [Jatropha curcas] |
| 8 |
Hb_002836_140 |
0.0989949522 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 9 |
Hb_000915_080 |
0.1015384445 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 10 |
Hb_000136_110 |
0.1071435361 |
- |
- |
hypothetical protein B456_011G031000, partial [Gossypium raimondii] |
| 11 |
Hb_000959_230 |
0.1076897404 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 12 homolog A [Populus euphratica] |
| 12 |
Hb_027472_120 |
0.1090770076 |
- |
- |
PREDICTED: thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial isoform X1 [Jatropha curcas] |
| 13 |
Hb_044662_060 |
0.1092242099 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 9, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000556_150 |
0.1107093643 |
- |
- |
PREDICTED: fasciclin-like arabinogalactan protein 10 [Populus euphratica] |
| 15 |
Hb_002498_140 |
0.1112266562 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22-like isoform X1 [Solanum lycopersicum] |
| 16 |
Hb_000487_180 |
0.1112883048 |
- |
- |
PREDICTED: post-GPI attachment to proteins factor 3 isoform X1 [Jatropha curcas] |
| 17 |
Hb_002351_030 |
0.1118100749 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_000414_050 |
0.1122612549 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 19 |
Hb_004007_220 |
0.1136994823 |
- |
- |
PREDICTED: casein kinase II subunit alpha [Jatropha curcas] |
| 20 |
Hb_058999_030 |
0.1141840045 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |