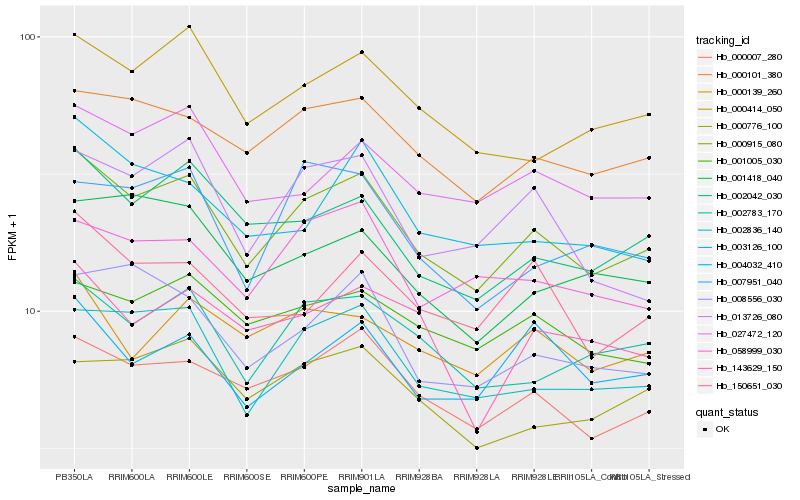
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000915_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628777 [Jatropha curcas] |
| 2 |
Hb_000007_280 |
0.0628084703 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 3 |
Hb_002836_140 |
0.062836661 |
- |
- |
PREDICTED: uncharacterized protein C1450.15 [Jatropha curcas] |
| 4 |
Hb_000101_380 |
0.067135195 |
- |
- |
PREDICTED: clathrin light chain 2-like isoform X2 [Populus euphratica] |
| 5 |
Hb_002042_030 |
0.0710722972 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 6 |
Hb_002783_170 |
0.0716126588 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X2 [Jatropha curcas] |
| 7 |
Hb_000414_050 |
0.073540958 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 8 |
Hb_027472_120 |
0.0768515506 |
- |
- |
PREDICTED: thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_008556_030 |
0.080851271 |
- |
- |
PREDICTED: uncharacterized protein LOC105648021 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001005_030 |
0.0823778374 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 11 |
Hb_150651_030 |
0.0824417044 |
- |
- |
PREDICTED: metallophosphoesterase 1-like [Jatropha curcas] |
| 12 |
Hb_058999_030 |
0.0842053506 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 13 |
Hb_001418_040 |
0.084329491 |
- |
- |
PREDICTED: mannosyl-oligosaccharide 1,2-alpha-mannosidase MNS1-like [Jatropha curcas] |
| 14 |
Hb_013726_080 |
0.085204768 |
- |
- |
Glycogen synthase kinase-3 beta, putative [Ricinus communis] |
| 15 |
Hb_000776_100 |
0.085219184 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 16 |
Hb_003126_100 |
0.0865877085 |
- |
- |
hypothetical protein POPTR_0005s11280g [Populus trichocarpa] |
| 17 |
Hb_004032_410 |
0.0867831526 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 18 |
Hb_007951_040 |
0.0869539685 |
- |
- |
signal peptidase family protein [Populus trichocarpa] |
| 19 |
Hb_000139_260 |
0.0874503754 |
- |
- |
PREDICTED: protein phosphatase 2C 70 [Jatropha curcas] |
| 20 |
Hb_143629_150 |
0.0875847861 |
- |
- |
PREDICTED: flap endonuclease 1 isoform X2 [Prunus mume] |