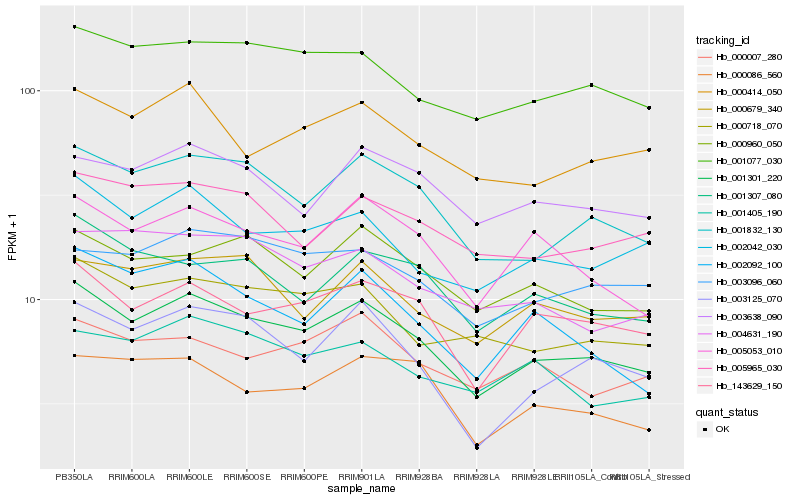
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001301_220 |
0.0 |
- |
- |
PREDICTED: polyadenylate-binding protein-interacting protein 4 [Jatropha curcas] |
| 2 |
Hb_001832_130 |
0.0585445735 |
- |
- |
PREDICTED: WD-40 repeat-containing protein MSI4 [Jatropha curcas] |
| 3 |
Hb_143629_150 |
0.0711428209 |
- |
- |
PREDICTED: flap endonuclease 1 isoform X2 [Prunus mume] |
| 4 |
Hb_001077_030 |
0.0738516152 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_002042_030 |
0.0748624489 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |
| 6 |
Hb_005053_010 |
0.0769075888 |
- |
- |
PREDICTED: protease Do-like 9 [Jatropha curcas] |
| 7 |
Hb_000718_070 |
0.0778541628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000960_050 |
0.0801062114 |
- |
- |
PREDICTED: protein EXPORTIN 1A [Jatropha curcas] |
| 9 |
Hb_003125_070 |
0.0804528795 |
- |
- |
PREDICTED: serine/threonine-protein kinase rio1-like [Jatropha curcas] |
| 10 |
Hb_001307_080 |
0.0819338427 |
- |
- |
PREDICTED: phospholipid--sterol O-acyltransferase [Jatropha curcas] |
| 11 |
Hb_000414_050 |
0.0829258073 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 12 |
Hb_000679_340 |
0.0844935873 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003638_090 |
0.0858219849 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 14 |
Hb_005965_030 |
0.0863624163 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 15 |
Hb_003096_060 |
0.086585974 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 16 |
Hb_002092_100 |
0.0870149843 |
- |
- |
PREDICTED: actin-related protein 4-like [Jatropha curcas] |
| 17 |
Hb_000086_560 |
0.0871449411 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase SUVR2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001405_190 |
0.0880414606 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 19 |
Hb_000007_280 |
0.0881533898 |
- |
- |
hypothetical protein VITISV_000066 [Vitis vinifera] |
| 20 |
Hb_004631_190 |
0.0887963752 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |