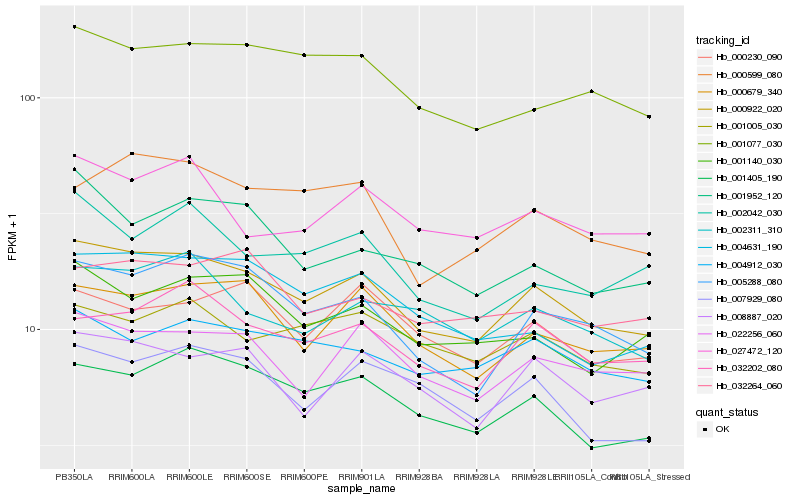
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_020 |
0.0 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 2 |
Hb_001405_190 |
0.0641416498 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_005288_080 |
0.0656445704 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000679_340 |
0.0673996818 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 5 |
Hb_022256_060 |
0.0704188741 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 6 |
Hb_032202_080 |
0.071266909 |
- |
- |
PREDICTED: peroxisome biogenesis protein 16-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_001077_030 |
0.0734050177 |
- |
- |
PREDICTED: somatic embryogenesis receptor kinase 1 isoform X2 [Jatropha curcas] |
| 8 |
Hb_008887_020 |
0.0738858907 |
- |
- |
PREDICTED: uncharacterized protein LOC105636191 [Jatropha curcas] |
| 9 |
Hb_004912_030 |
0.0750238601 |
transcription factor |
TF Family: Jumonji |
PREDICTED: uncharacterized protein LOC105650086 [Jatropha curcas] |
| 10 |
Hb_007929_080 |
0.0752623344 |
- |
- |
PREDICTED: transforming growth factor-beta receptor-associated protein 1 [Jatropha curcas] |
| 11 |
Hb_002311_310 |
0.0755570574 |
- |
- |
PREDICTED: uncharacterized protein LOC100246622 [Vitis vinifera] |
| 12 |
Hb_004631_190 |
0.0767294027 |
- |
- |
PREDICTED: protein SCAI homolog [Jatropha curcas] |
| 13 |
Hb_000599_080 |
0.0770018971 |
- |
- |
PREDICTED: TOM1-like protein 1 [Malus domestica] |
| 14 |
Hb_001140_030 |
0.0783571568 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase XBAT33 [Jatropha curcas] |
| 15 |
Hb_000230_090 |
0.0807219667 |
- |
- |
PREDICTED: WD repeat-containing protein 48 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001005_030 |
0.0807897671 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH4 isoform X3 [Jatropha curcas] |
| 17 |
Hb_027472_120 |
0.0811554338 |
- |
- |
PREDICTED: thiosulfate/3-mercaptopyruvate sulfurtransferase 1, mitochondrial isoform X1 [Jatropha curcas] |
| 18 |
Hb_032264_060 |
0.0819126743 |
- |
- |
hypothetical protein JCGZ_02506 [Jatropha curcas] |
| 19 |
Hb_001952_120 |
0.0823093865 |
- |
- |
PREDICTED: polyadenylate-binding protein 2-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_002042_030 |
0.0831361025 |
- |
- |
PREDICTED: protein YIPF1 homolog [Jatropha curcas] |