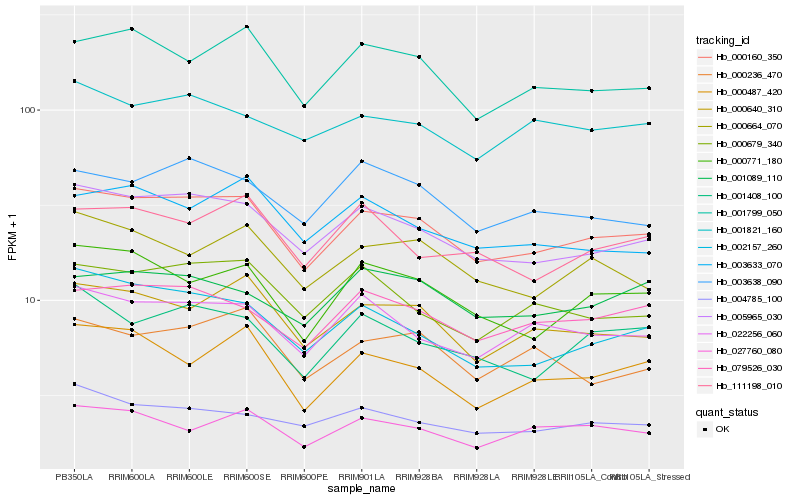
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000160_350 |
0.0 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 4 isoform X3 [Vitis vinifera] |
| 2 |
Hb_005965_030 |
0.0422792461 |
- |
- |
hypothetical protein JCGZ_16415 [Jatropha curcas] |
| 3 |
Hb_000640_310 |
0.060451158 |
- |
- |
hypothetical protein JCGZ_14055 [Jatropha curcas] |
| 4 |
Hb_001799_050 |
0.0635226103 |
- |
- |
PREDICTED: plasminogen activator inhibitor 1 RNA-binding protein [Vitis vinifera] |
| 5 |
Hb_079526_030 |
0.0638407579 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 6 |
Hb_000771_180 |
0.0649669783 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 7 |
Hb_022256_060 |
0.0669186448 |
- |
- |
PREDICTED: uncharacterized protein LOC105628137 [Jatropha curcas] |
| 8 |
Hb_003638_090 |
0.0712040963 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor SR45a-like [Jatropha curcas] |
| 9 |
Hb_000664_070 |
0.0715672783 |
- |
- |
PREDICTED: protein cereblon [Jatropha curcas] |
| 10 |
Hb_000679_340 |
0.0720699751 |
- |
- |
PREDICTED: uncharacterized protein LOC105650070 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002157_260 |
0.0733302645 |
- |
- |
PREDICTED: partner of Y14 and mago-like [Jatropha curcas] |
| 12 |
Hb_004785_100 |
0.0737656747 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001408_100 |
0.0756659918 |
- |
- |
pseudouridylate synthase, putative [Ricinus communis] |
| 14 |
Hb_001821_160 |
0.076029585 |
- |
- |
serine/arginine rich splicing factor, putative [Ricinus communis] |
| 15 |
Hb_027760_080 |
0.0768255591 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_003633_070 |
0.0770497193 |
- |
- |
PREDICTED: uncharacterized protein LOC105647493 [Jatropha curcas] |
| 17 |
Hb_001089_110 |
0.0779113372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000487_420 |
0.0790494538 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At1g04390 [Jatropha curcas] |
| 19 |
Hb_000236_470 |
0.0797212789 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g55840 [Jatropha curcas] |
| 20 |
Hb_111198_010 |
0.0798512791 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 37-like isoform X1 [Jatropha curcas] |