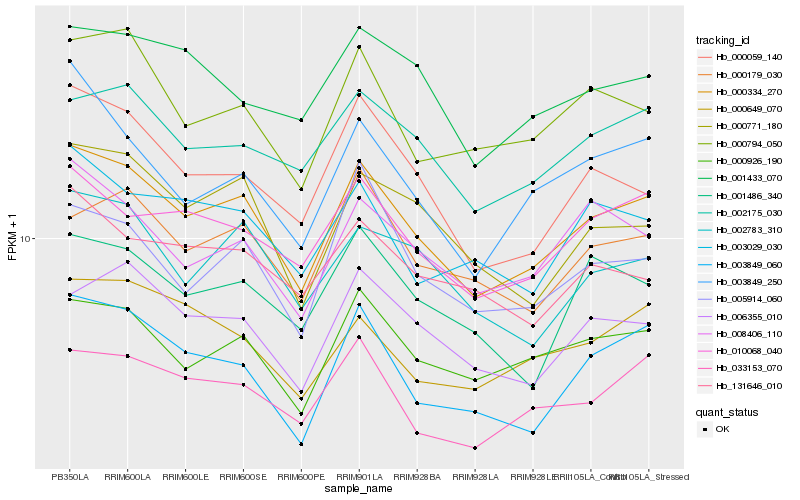
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000334_270 |
0.0 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000926_190 |
0.0596963465 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 3, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 3 |
Hb_010068_040 |
0.0623512892 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |
| 4 |
Hb_003849_060 |
0.0635983557 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g13040, mitochondrial [Jatropha curcas] |
| 5 |
Hb_033153_070 |
0.064889978 |
- |
- |
PREDICTED: uncharacterized protein LOC105649606 isoform X1 [Jatropha curcas] |
| 6 |
Hb_006355_010 |
0.0657128303 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 7 |
Hb_002175_030 |
0.065918619 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 8 |
Hb_000771_180 |
0.0664312737 |
- |
- |
polypyrimidine tract binding protein, putative [Ricinus communis] |
| 9 |
Hb_000059_140 |
0.066484639 |
- |
- |
rnase h, putative [Ricinus communis] |
| 10 |
Hb_008406_110 |
0.0669761116 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RING1a isoform X1 [Jatropha curcas] |
| 11 |
Hb_000794_050 |
0.067863811 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 12 [Jatropha curcas] |
| 12 |
Hb_002783_310 |
0.0682416937 |
- |
- |
PREDICTED: tRNA (adenine(58)-N(1))-methyltransferase non-catalytic subunit trm6 [Jatropha curcas] |
| 13 |
Hb_000179_030 |
0.070451638 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000649_070 |
0.072127835 |
- |
- |
PREDICTED: probable UDP-3-O-[3-hydroxymyristoyl] N-acetylglucosamine deacetylase 2 [Jatropha curcas] |
| 15 |
Hb_003029_030 |
0.0721781487 |
- |
- |
PREDICTED: uncharacterized protein LOC105642632 isoform X2 [Jatropha curcas] |
| 16 |
Hb_003849_250 |
0.0727536671 |
- |
- |
PREDICTED: protein MAK16 homolog [Jatropha curcas] |
| 17 |
Hb_005914_060 |
0.0738076235 |
- |
- |
PREDICTED: splicing factor U2af large subunit A [Jatropha curcas] |
| 18 |
Hb_001486_340 |
0.0743510127 |
- |
- |
PREDICTED: double-stranded RNA-binding protein 1-like [Jatropha curcas] |
| 19 |
Hb_001433_070 |
0.0750613422 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 20 |
Hb_131646_010 |
0.0754293257 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |