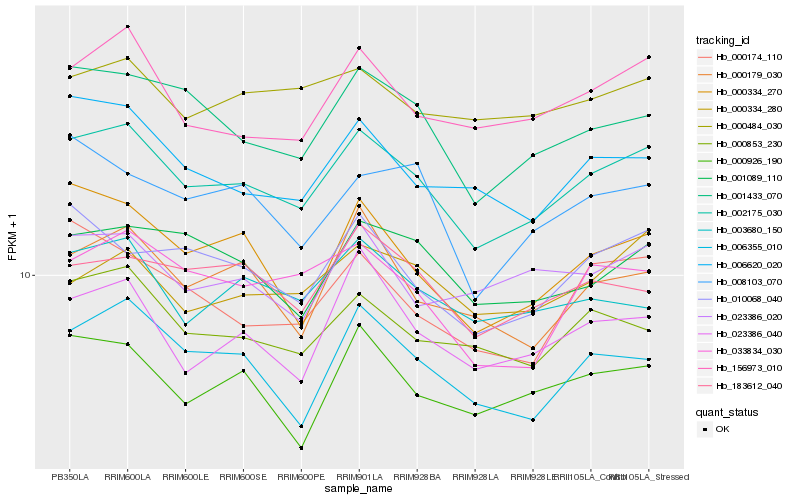
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002175_030 |
0.0 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 2 |
Hb_001433_070 |
0.0559059494 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 3 |
Hb_156973_010 |
0.0581205892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000853_230 |
0.0609517788 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 5 |
Hb_001089_110 |
0.0640989079 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_010068_040 |
0.0643016809 |
- |
- |
PREDICTED: uncharacterized protein LOC105632964 [Jatropha curcas] |
| 7 |
Hb_006620_020 |
0.0649246383 |
- |
- |
PREDICTED: uncharacterized protein LOC105638878 [Jatropha curcas] |
| 8 |
Hb_006355_010 |
0.06532327 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 9 |
Hb_033834_030 |
0.0658516574 |
- |
- |
PREDICTED: nucleolar complex protein 2 homolog isoform X1 [Jatropha curcas] |
| 10 |
Hb_000334_270 |
0.065918619 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 11 |
Hb_183612_040 |
0.0676410475 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 12 |
Hb_000334_280 |
0.0687991914 |
- |
- |
PREDICTED: protein FRA10AC1 [Jatropha curcas] |
| 13 |
Hb_000926_190 |
0.0687996603 |
- |
- |
PREDICTED: 1,4-alpha-glucan-branching enzyme 3, chloroplastic/amyloplastic isoform X2 [Jatropha curcas] |
| 14 |
Hb_000179_030 |
0.0690827397 |
- |
- |
PREDICTED: zinc finger protein VAR3, chloroplastic [Jatropha curcas] |
| 15 |
Hb_003680_150 |
0.0699043993 |
- |
- |
srpk, putative [Ricinus communis] |
| 16 |
Hb_023386_040 |
0.070568759 |
- |
- |
PREDICTED: chaperone protein dnaJ 13-like [Populus euphratica] |
| 17 |
Hb_000174_110 |
0.0706792034 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 18 |
Hb_023386_020 |
0.0706951019 |
- |
- |
PREDICTED: chaperone protein dnaJ 13 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000484_030 |
0.0710331888 |
- |
- |
PREDICTED: probable 26S proteasome non-ATPase regulatory subunit 3 [Jatropha curcas] |
| 20 |
Hb_008103_070 |
0.0716249854 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |