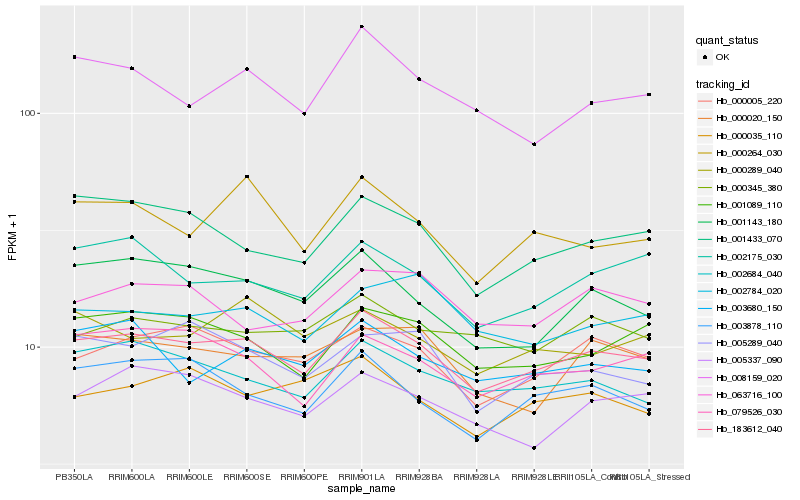
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_183612_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 2 |
Hb_000005_220 |
0.0570896652 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 3 |
Hb_001089_110 |
0.0594251364 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002684_040 |
0.0595959601 |
- |
- |
PREDICTED: uncharacterized protein LOC105634392 isoform X1 [Jatropha curcas] |
| 5 |
Hb_001433_070 |
0.0648713221 |
- |
- |
PREDICTED: splicing regulatory glutamine/lysine-rich protein 1 [Jatropha curcas] |
| 6 |
Hb_003878_110 |
0.0654276316 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 7 |
Hb_079526_030 |
0.0660309737 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 8 |
Hb_001143_180 |
0.0672857725 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 9 |
Hb_002175_030 |
0.0676410475 |
- |
- |
PREDICTED: snRNA-activating protein complex subunit isoform X2 [Jatropha curcas] |
| 10 |
Hb_000345_380 |
0.0682698612 |
- |
- |
PREDICTED: long chain acyl-CoA synthetase 7, peroxisomal [Jatropha curcas] |
| 11 |
Hb_005337_090 |
0.0683562729 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 12 |
Hb_003680_150 |
0.068492241 |
- |
- |
srpk, putative [Ricinus communis] |
| 13 |
Hb_008159_020 |
0.0691294961 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit beta-like protein [Jatropha curcas] |
| 14 |
Hb_000264_030 |
0.0697692151 |
- |
- |
PREDICTED: splicing factor 3B subunit 2 [Jatropha curcas] |
| 15 |
Hb_005289_040 |
0.071652109 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 16 |
Hb_002784_020 |
0.0731505337 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 3 [Jatropha curcas] |
| 17 |
Hb_000020_150 |
0.0733292002 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |
| 18 |
Hb_000035_110 |
0.0735638909 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 19 |
Hb_063716_100 |
0.0736371232 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 56-like isoform X2 [Jatropha curcas] |
| 20 |
Hb_000289_040 |
0.0736422421 |
- |
- |
Poly(A) polymerase alpha, putative [Ricinus communis] |