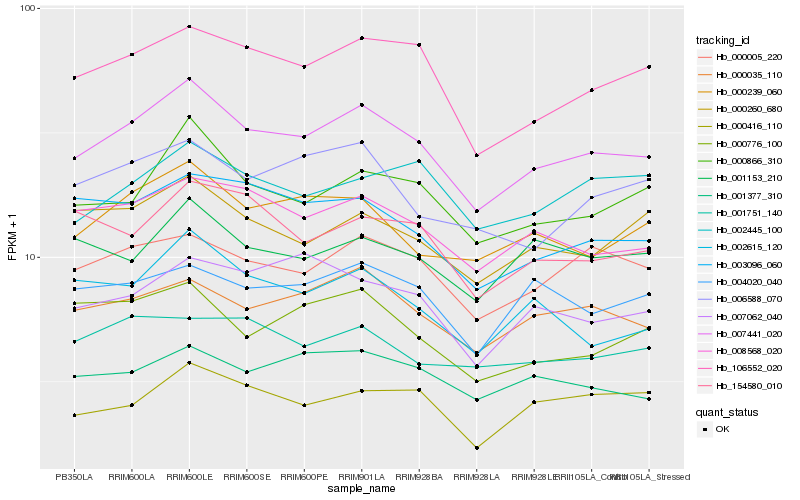
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_020 |
0.0 |
- |
- |
Rab6 [Hevea brasiliensis] |
| 2 |
Hb_106552_020 |
0.060043012 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000866_310 |
0.0625589318 |
- |
- |
PREDICTED: neural Wiskott-Aldrich syndrome protein [Jatropha curcas] |
| 4 |
Hb_000035_110 |
0.0648206015 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 5 [Jatropha curcas] |
| 5 |
Hb_008568_020 |
0.0653888017 |
- |
- |
PREDICTED: uncharacterized protein LOC105122325 isoform X3 [Populus euphratica] |
| 6 |
Hb_000005_220 |
0.0658703319 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 7 |
Hb_000239_060 |
0.0695997778 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_003096_060 |
0.0739550196 |
- |
- |
PREDICTED: LMBR1 domain-containing protein 2 homolog A [Jatropha curcas] |
| 9 |
Hb_000416_110 |
0.0754435283 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase RFK1 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001377_310 |
0.0757984435 |
transcription factor |
TF Family: Coactivator p15 |
PREDICTED: uncharacterized protein LOC105642839 [Jatropha curcas] |
| 11 |
Hb_000260_680 |
0.0761562792 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 12 |
Hb_006588_070 |
0.0779692942 |
- |
- |
20 kD nuclear cap binding protein, putative [Ricinus communis] |
| 13 |
Hb_004020_040 |
0.0781169419 |
- |
- |
PREDICTED: GTP-binding protein At3g49725, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002615_120 |
0.079671235 |
- |
- |
PREDICTED: rho GTPase-activating protein 7 isoform X1 [Jatropha curcas] |
| 15 |
Hb_154580_010 |
0.0799756293 |
- |
- |
hypothetical protein PRUPE_ppa002109mg [Prunus persica] |
| 16 |
Hb_002445_100 |
0.0809580345 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000776_100 |
0.0810021296 |
- |
- |
Cleavage stimulation factor 50 kDa subunit, putative [Ricinus communis] |
| 18 |
Hb_001153_210 |
0.0816341494 |
- |
- |
PREDICTED: uncharacterized protein LOC105645887 isoform X1 [Jatropha curcas] |
| 19 |
Hb_007062_040 |
0.0821218934 |
- |
- |
ADP-ribosylation factor GTPase-activating protein AGD2 [Morus notabilis] |
| 20 |
Hb_001751_140 |
0.0826054892 |
- |
- |
PREDICTED: uncharacterized protein LOC105648510 isoform X1 [Jatropha curcas] |