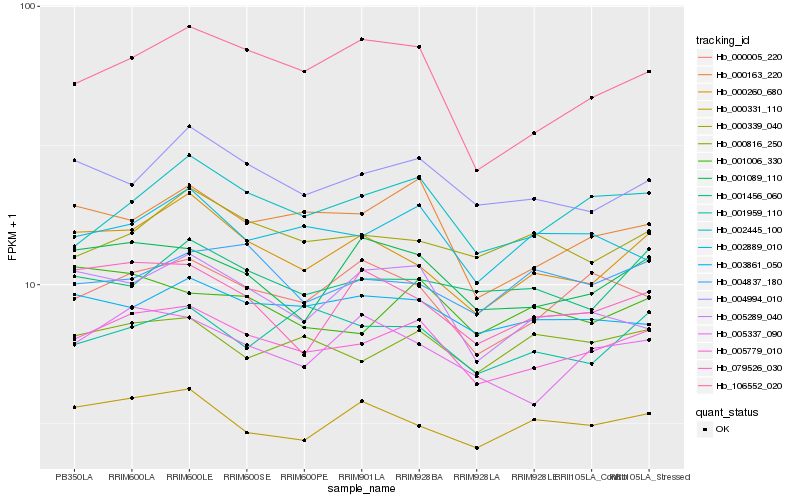
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005779_010 |
0.0 |
- |
- |
catalytic, putative [Ricinus communis] |
| 2 |
Hb_004994_010 |
0.0626880836 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 1 [Jatropha curcas] |
| 3 |
Hb_002445_100 |
0.0630142704 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 4 |
Hb_000163_220 |
0.0630555432 |
- |
- |
hypothetical protein CISIN_1g022301mg [Citrus sinensis] |
| 5 |
Hb_000816_250 |
0.0651245869 |
- |
- |
PREDICTED: uncharacterized protein LOC105649197 isoform X1 [Jatropha curcas] |
| 6 |
Hb_003861_050 |
0.0656288966 |
- |
- |
PREDICTED: uncharacterized protein LOC105650779 [Jatropha curcas] |
| 7 |
Hb_000005_220 |
0.0672703747 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 8 |
Hb_000260_680 |
0.0673555516 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 9 |
Hb_002889_010 |
0.0674454223 |
- |
- |
ubx domain-containing, putative [Ricinus communis] |
| 10 |
Hb_005337_090 |
0.0686673127 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 11 |
Hb_000331_110 |
0.0692451764 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 12 |
Hb_001089_110 |
0.0698488254 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_106552_020 |
0.071346021 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RS2Z33 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000339_040 |
0.0735208025 |
- |
- |
PREDICTED: SAP30-binding protein isoform X2 [Jatropha curcas] |
| 15 |
Hb_001006_330 |
0.0737244003 |
- |
- |
sodium/hydrogen exchanger plant, putative [Ricinus communis] |
| 16 |
Hb_001456_060 |
0.0741343689 |
- |
- |
hypothetical protein JCGZ_17090 [Jatropha curcas] |
| 17 |
Hb_001959_110 |
0.0751702096 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 18 |
Hb_005289_040 |
0.0762834689 |
- |
- |
poly(A) polymerase, putative [Ricinus communis] |
| 19 |
Hb_079526_030 |
0.0769600446 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 20 |
Hb_004837_180 |
0.0786076662 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |