| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
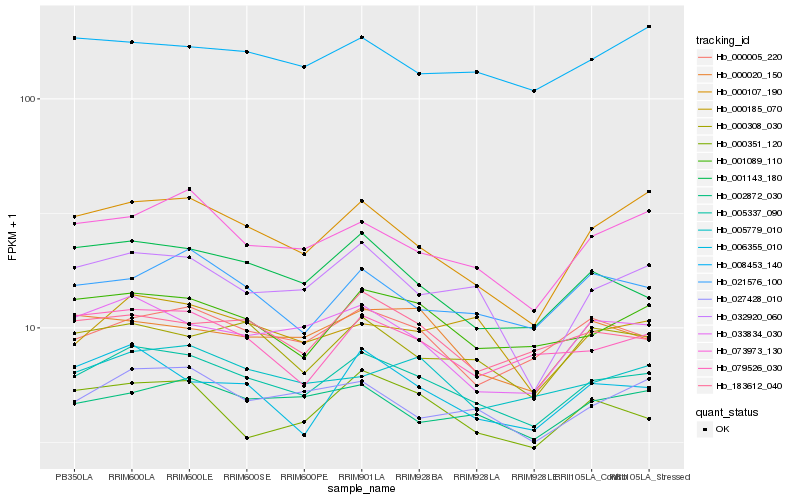
Hb_005337_090 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 2 |
Hb_001089_110 |
0.0576195926 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_027428_010 |
0.0626408161 |
- |
- |
alpha-1,3-mannosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_000308_030 |
0.0637927854 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 5 |
Hb_000005_220 |
0.0640651849 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 6 |
Hb_073973_130 |
0.0647566726 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |
| 7 |
Hb_033834_030 |
0.0655613279 |
- |
- |
PREDICTED: nucleolar complex protein 2 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_002872_030 |
0.0669280199 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 9 |
Hb_001143_180 |
0.0678503253 |
- |
- |
PREDICTED: replication factor C subunit 5 [Jatropha curcas] |
| 10 |
Hb_183612_040 |
0.0683562729 |
- |
- |
PREDICTED: uncharacterized protein LOC105632370 [Jatropha curcas] |
| 11 |
Hb_005779_010 |
0.0686673127 |
- |
- |
catalytic, putative [Ricinus communis] |
| 12 |
Hb_006355_010 |
0.06880387 |
- |
- |
PREDICTED: protein NLRC3 [Jatropha curcas] |
| 13 |
Hb_021576_100 |
0.0691022611 |
- |
- |
Enoyl-CoA hydratase, mitochondrial precursor, putative [Ricinus communis] |
| 14 |
Hb_079526_030 |
0.0692819347 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 15 |
Hb_000185_070 |
0.0703352903 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000351_120 |
0.0711427068 |
- |
- |
hypothetical protein POPTR_0013s10290g [Populus trichocarpa] |
| 17 |
Hb_032920_060 |
0.0722005568 |
- |
- |
PREDICTED: mitotic checkpoint protein BUB3.1 [Jatropha curcas] |
| 18 |
Hb_008453_140 |
0.0729459166 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 19 |
Hb_000107_190 |
0.0730727349 |
- |
- |
50S ribosomal protein L20 [Hevea brasiliensis] |
| 20 |
Hb_000020_150 |
0.0734932038 |
- |
- |
PREDICTED: uncharacterized protein LOC105636325 [Jatropha curcas] |