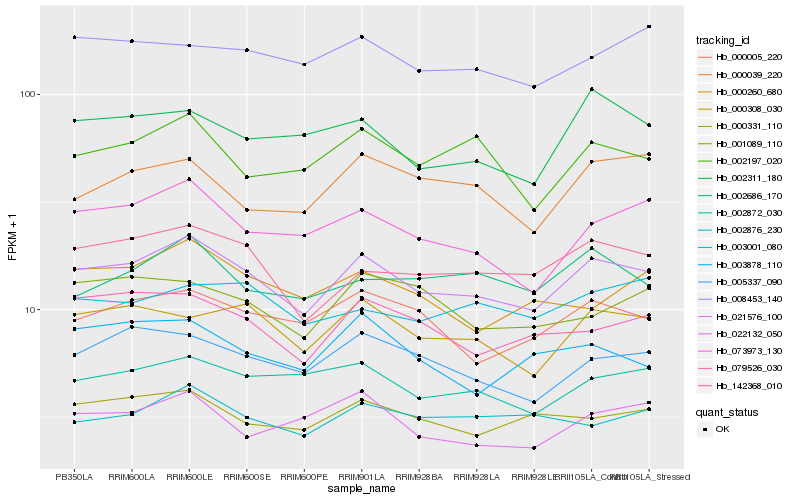
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021576_100 |
0.0 |
- |
- |
Enoyl-CoA hydratase, mitochondrial precursor, putative [Ricinus communis] |
| 2 |
Hb_142368_010 |
0.0665533799 |
- |
- |
mRNA (guanine-7-)methyltransferase, putative [Ricinus communis] |
| 3 |
Hb_000308_030 |
0.0689905127 |
- |
- |
PREDICTED: uncharacterized protein LOC105639686 [Jatropha curcas] |
| 4 |
Hb_005337_090 |
0.0691022611 |
- |
- |
PREDICTED: serine/threonine-protein kinase TOUSLED-like [Jatropha curcas] |
| 5 |
Hb_073973_130 |
0.0705314857 |
- |
- |
PREDICTED: prolyl-tRNA synthetase associated domain-containing protein 1 [Jatropha curcas] |
| 6 |
Hb_000005_220 |
0.071629537 |
- |
- |
DNA ligase 1, partial [Glycine soja] |
| 7 |
Hb_079526_030 |
0.073472509 |
- |
- |
PREDICTED: uncharacterized protein LOC105638120 [Jatropha curcas] |
| 8 |
Hb_002872_030 |
0.0739796205 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |
| 9 |
Hb_000331_110 |
0.0741729302 |
- |
- |
Conserved oligomeric Golgi complex component, putative [Ricinus communis] |
| 10 |
Hb_002197_020 |
0.0779221002 |
- |
- |
DNA-directed RNA polymerase family protein [Theobroma cacao] |
| 11 |
Hb_002311_180 |
0.0801203771 |
- |
- |
drought-responsive family protein [Populus trichocarpa] |
| 12 |
Hb_002876_230 |
0.0814623527 |
- |
- |
PREDICTED: uncharacterized protein LOC105633933 [Jatropha curcas] |
| 13 |
Hb_002686_170 |
0.0826731689 |
- |
- |
PREDICTED: uncharacterized protein LOC105632645 isoform X1 [Jatropha curcas] |
| 14 |
Hb_008453_140 |
0.0828232652 |
- |
- |
PREDICTED: SKP1-like protein 1B [Jatropha curcas] |
| 15 |
Hb_001089_110 |
0.0832389085 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000039_220 |
0.0833643289 |
- |
- |
PREDICTED: vesicle-associated protein 4-2-like [Gossypium raimondii] |
| 17 |
Hb_022132_050 |
0.0845245429 |
- |
- |
GTP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000260_680 |
0.0849438223 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 4 regulatory subunit 2-A [Jatropha curcas] |
| 19 |
Hb_003878_110 |
0.0853012575 |
- |
- |
PREDICTED: uncharacterized protein LOC105637024 [Jatropha curcas] |
| 20 |
Hb_003001_080 |
0.0856606483 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |