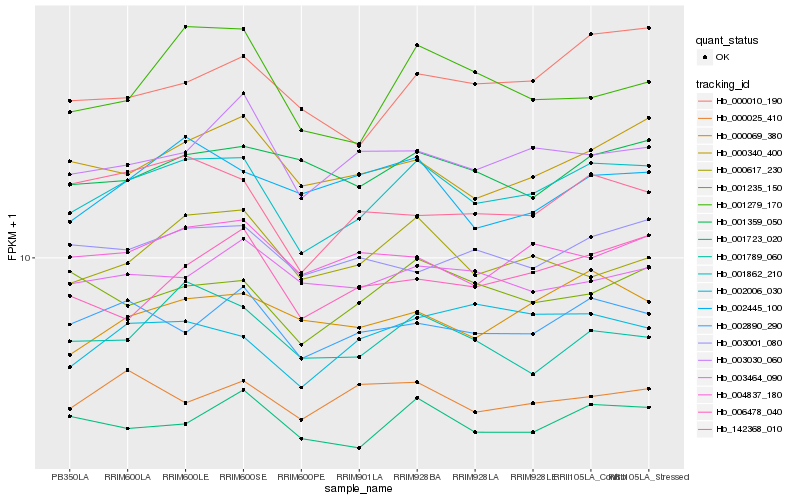
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001862_210 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: salutaridine reductase-like [Citrus sinensis] |
| 2 |
Hb_001723_020 |
0.0776530477 |
- |
- |
PREDICTED: nuclear export mediator factor Nemf [Jatropha curcas] |
| 3 |
Hb_002890_290 |
0.0793642146 |
- |
- |
PREDICTED: outer envelope protein 64, mitochondrial [Jatropha curcas] |
| 4 |
Hb_000340_400 |
0.0816681066 |
- |
- |
Histidine-containing phosphotransfer protein, putative [Ricinus communis] |
| 5 |
Hb_142368_010 |
0.0845706442 |
- |
- |
mRNA (guanine-7-)methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_004837_180 |
0.0853244241 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 7 |
Hb_000069_380 |
0.0870714656 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 8 |
Hb_000617_230 |
0.0876956955 |
- |
- |
PREDICTED: tafazzin isoform X1 [Jatropha curcas] |
| 9 |
Hb_001235_150 |
0.0878110078 |
- |
- |
PREDICTED: chaperone protein dnaJ 13 isoform X1 [Jatropha curcas] |
| 10 |
Hb_001789_060 |
0.0880791199 |
- |
- |
calmodulin-binding heat-shock protein, putative [Ricinus communis] |
| 11 |
Hb_003030_060 |
0.0881900727 |
- |
- |
PREDICTED: regulation of nuclear pre-mRNA domain-containing protein 2-like [Jatropha curcas] |
| 12 |
Hb_000010_190 |
0.0894933248 |
- |
- |
PREDICTED: protein-L-isoaspartate O-methyltransferase 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_003001_080 |
0.0895601957 |
- |
- |
PREDICTED: uncharacterized protein LOC105644363 [Jatropha curcas] |
| 14 |
Hb_006478_040 |
0.0899199611 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase subunit alpha, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001279_170 |
0.0912002501 |
- |
- |
PREDICTED: succinate dehydrogenase [ubiquinone] iron-sulfur subunit 2, mitochondrial [Jatropha curcas] |
| 16 |
Hb_002006_030 |
0.0921840607 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER [Jatropha curcas] |
| 17 |
Hb_001359_050 |
0.0928021929 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_002445_100 |
0.09288469 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003464_090 |
0.0932222621 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105642342 [Jatropha curcas] |
| 20 |
Hb_000025_410 |
0.094421091 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g01110-like [Populus euphratica] |