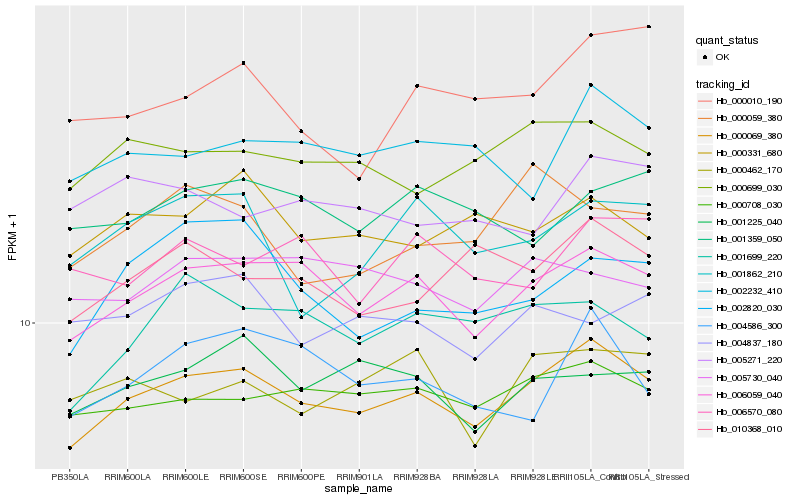
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000069_380 |
0.0 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 2 |
Hb_006059_040 |
0.0455037826 |
transcription factor |
TF Family: SOH1 |
hypothetical protein PHAVU_007G063000g [Phaseolus vulgaris] |
| 3 |
Hb_000708_030 |
0.0666834254 |
- |
- |
PREDICTED: uncharacterized protein LOC105650742 [Jatropha curcas] |
| 4 |
Hb_000699_030 |
0.0726038323 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 5 |
Hb_001225_040 |
0.0783949386 |
- |
- |
PREDICTED: meiosis-specific nuclear structural protein 1 isoform X5 [Jatropha curcas] |
| 6 |
Hb_010368_010 |
0.0785390243 |
- |
- |
RNA splicing protein mrs2, mitochondrial, putative [Ricinus communis] |
| 7 |
Hb_001699_220 |
0.0795118155 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH121-like [Jatropha curcas] |
| 8 |
Hb_002232_410 |
0.0802995966 |
- |
- |
PREDICTED: L-galactose dehydrogenase [Jatropha curcas] |
| 9 |
Hb_000331_680 |
0.0809813403 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 10 |
Hb_005730_040 |
0.0839212232 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 11 |
Hb_004586_300 |
0.0847375316 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000462_170 |
0.0850837068 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001359_050 |
0.0866719788 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001862_210 |
0.0870714656 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: salutaridine reductase-like [Citrus sinensis] |
| 15 |
Hb_002820_030 |
0.0884302659 |
- |
- |
PREDICTED: uncharacterized protein LOC105638922 [Jatropha curcas] |
| 16 |
Hb_000059_380 |
0.0887033969 |
- |
- |
PREDICTED: uncharacterized protein LOC105648668 [Jatropha curcas] |
| 17 |
Hb_000010_190 |
0.0889887877 |
- |
- |
PREDICTED: protein-L-isoaspartate O-methyltransferase 1 isoform X2 [Jatropha curcas] |
| 18 |
Hb_006570_080 |
0.0897959709 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |
| 19 |
Hb_004837_180 |
0.0901678262 |
transcription factor |
TF Family: NF-YA |
PREDICTED: nuclear transcription factor Y subunit A-1-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_005271_220 |
0.0902673903 |
- |
- |
PREDICTED: general transcription factor IIE subunit 2-like [Jatropha curcas] |