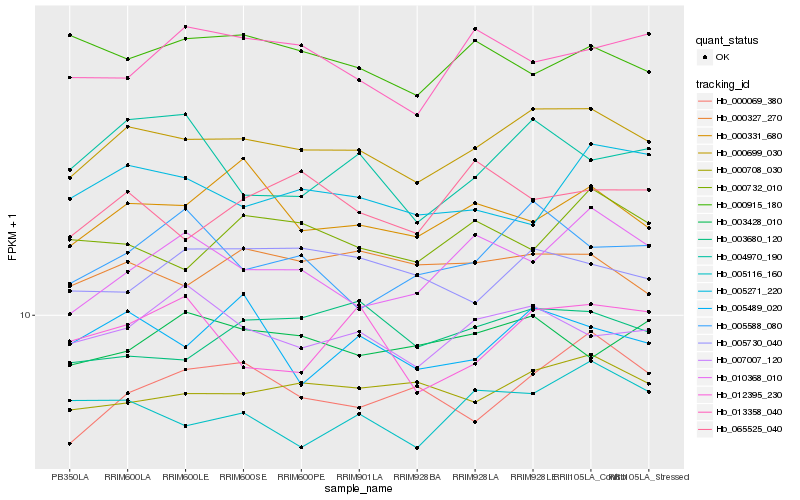
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000699_030 |
0.0 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 2 |
Hb_000708_030 |
0.0554491762 |
- |
- |
PREDICTED: uncharacterized protein LOC105650742 [Jatropha curcas] |
| 3 |
Hb_000327_270 |
0.064212994 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 4 |
Hb_007007_120 |
0.0645188786 |
- |
- |
PREDICTED: lysophospholipid acyltransferase LPEAT2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_005116_160 |
0.0676157615 |
transcription factor |
TF Family: TRAF |
- |
| 6 |
Hb_005489_020 |
0.0681678494 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_003680_120 |
0.0684811582 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase UPL7 [Jatropha curcas] |
| 8 |
Hb_000331_680 |
0.0689254866 |
- |
- |
Protein bem46, putative [Ricinus communis] |
| 9 |
Hb_005730_040 |
0.0716287775 |
- |
- |
hypothetical protein PRUPE_ppa004338mg [Prunus persica] |
| 10 |
Hb_003428_010 |
0.071656102 |
- |
- |
PREDICTED: probable NAD(P)H dehydrogenase (quinone) FQR1-like 1 [Jatropha curcas] |
| 11 |
Hb_005588_080 |
0.0722040709 |
- |
- |
PREDICTED: uncharacterized protein LOC105633661 [Jatropha curcas] |
| 12 |
Hb_000069_380 |
0.0726038323 |
transcription factor |
TF Family: PHD |
PREDICTED: histone-lysine N-methyltransferase ATX3 isoform X2 [Jatropha curcas] |
| 13 |
Hb_010368_010 |
0.0727057181 |
- |
- |
RNA splicing protein mrs2, mitochondrial, putative [Ricinus communis] |
| 14 |
Hb_000732_010 |
0.0741381671 |
- |
- |
PREDICTED: nitric oxide synthase-interacting protein [Pyrus x bretschneideri] |
| 15 |
Hb_013358_040 |
0.0762527128 |
- |
- |
PREDICTED: stromal cell-derived factor 2-like protein [Jatropha curcas] |
| 16 |
Hb_065525_040 |
0.0767347178 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor 3-like [Jatropha curcas] |
| 17 |
Hb_004970_190 |
0.0772015758 |
- |
- |
DCL protein, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_005271_220 |
0.0782322551 |
- |
- |
PREDICTED: general transcription factor IIE subunit 2-like [Jatropha curcas] |
| 19 |
Hb_012395_230 |
0.0798148137 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 20 |
Hb_000915_180 |
0.0798791686 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Pyrus x bretschneideri] |