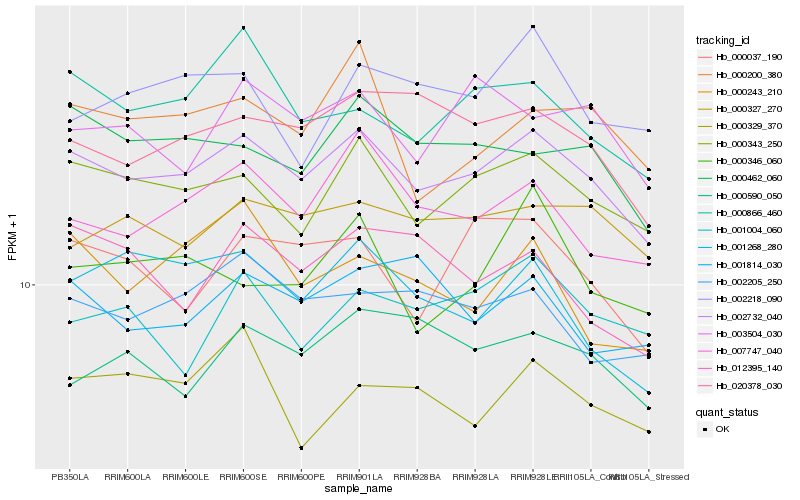
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002732_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000343_250 |
0.0415607474 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 3 |
Hb_007747_040 |
0.052805163 |
- |
- |
PREDICTED: probable UDP-N-acetylglucosamine--peptide N-acetylglucosaminyltransferase SPINDLY [Jatropha curcas] |
| 4 |
Hb_000462_060 |
0.0533741153 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105642316 isoform X2 [Jatropha curcas] |
| 5 |
Hb_020378_030 |
0.0547598984 |
- |
- |
PREDICTED: protein transport protein SEC31 homolog B [Jatropha curcas] |
| 6 |
Hb_000866_460 |
0.0559744956 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000327_270 |
0.0561245368 |
- |
- |
PREDICTED: leukocyte receptor cluster member 8 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_002205_250 |
0.0573498556 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000037_190 |
0.0617300019 |
- |
- |
PREDICTED: uncharacterized protein LOC105641500 [Jatropha curcas] |
| 10 |
Hb_000200_380 |
0.0622878926 |
- |
- |
PREDICTED: uncharacterized protein LOC105636933 isoform X2 [Jatropha curcas] |
| 11 |
Hb_003504_030 |
0.0627641447 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase PP1-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_001268_280 |
0.0642103171 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001814_030 |
0.066180541 |
- |
- |
PREDICTED: uncharacterized protein LOC105650634 [Jatropha curcas] |
| 14 |
Hb_000590_050 |
0.0662058029 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase MKP1 [Jatropha curcas] |
| 15 |
Hb_000329_370 |
0.067516604 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 16 |
Hb_012395_140 |
0.0676082837 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |
| 17 |
Hb_001004_060 |
0.0683014961 |
- |
- |
PREDICTED: uncharacterized protein LOC105630220 isoform X4 [Jatropha curcas] |
| 18 |
Hb_002218_090 |
0.0683298054 |
- |
- |
Poly(rC)-binding protein, putative [Ricinus communis] |
| 19 |
Hb_000346_060 |
0.0687451314 |
- |
- |
hypothetical protein JCGZ_19590 [Jatropha curcas] |
| 20 |
Hb_000243_210 |
0.0691135808 |
- |
- |
protein with unknown function [Ricinus communis] |