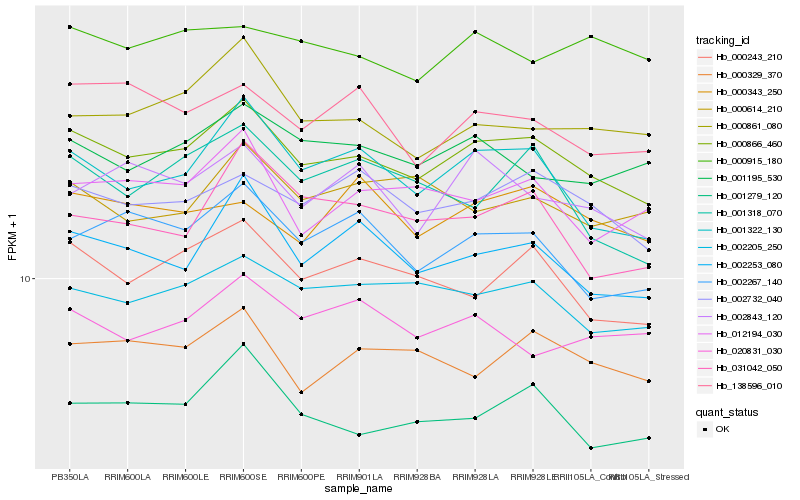
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000866_460 |
0.0 |
- |
- |
PREDICTED: acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_001322_130 |
0.0533215794 |
- |
- |
PREDICTED: putative glycerol-3-phosphate transporter 4 [Jatropha curcas] |
| 3 |
Hb_002205_250 |
0.0550337989 |
- |
- |
PREDICTED: probable inactive serine/threonine-protein kinase scy1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002732_040 |
0.0559744956 |
- |
- |
PREDICTED: uncharacterized protein LOC105628163 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000243_210 |
0.058064968 |
- |
- |
protein with unknown function [Ricinus communis] |
| 6 |
Hb_002253_080 |
0.0588162049 |
- |
- |
PREDICTED: phosphatidylinositol 4-kinase beta 1-like [Jatropha curcas] |
| 7 |
Hb_001279_120 |
0.0633058956 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Jatropha curcas] |
| 8 |
Hb_002267_140 |
0.0641614691 |
- |
- |
PREDICTED: VHS domain-containing protein At3g16270 [Jatropha curcas] |
| 9 |
Hb_001195_530 |
0.064245255 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000861_080 |
0.0645631744 |
- |
- |
PREDICTED: protein ELC-like [Jatropha curcas] |
| 11 |
Hb_000329_370 |
0.0652752007 |
- |
- |
hypothetical protein JCGZ_14571 [Jatropha curcas] |
| 12 |
Hb_012194_030 |
0.0657129008 |
- |
- |
PREDICTED: R3H domain-containing protein 2-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_138596_010 |
0.0675026556 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At2g13690 [Jatropha curcas] |
| 14 |
Hb_002843_120 |
0.0676680784 |
- |
- |
PREDICTED: uncharacterized protein LOC105645113 isoform X1 [Jatropha curcas] |
| 15 |
Hb_031042_050 |
0.0704212072 |
- |
- |
PREDICTED: protein transport protein SEC16B homolog [Jatropha curcas] |
| 16 |
Hb_000915_180 |
0.0715633705 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit-like [Pyrus x bretschneideri] |
| 17 |
Hb_000343_250 |
0.0724753352 |
- |
- |
PREDICTED: protein CASC3 [Jatropha curcas] |
| 18 |
Hb_001318_070 |
0.0728699766 |
transcription factor |
TF Family: SBP |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_020831_030 |
0.0729751066 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000614_210 |
0.0730471548 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-1-like isoform X1 [Jatropha curcas] |